Multiplexed detection of anti-red cell alloantibodies
A technology of alloantibodies and red blood cells, applied in measuring devices, immunoassays, biological tests, etc., can solve problems such as the difficulty of constructing a large number of unique passwords
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
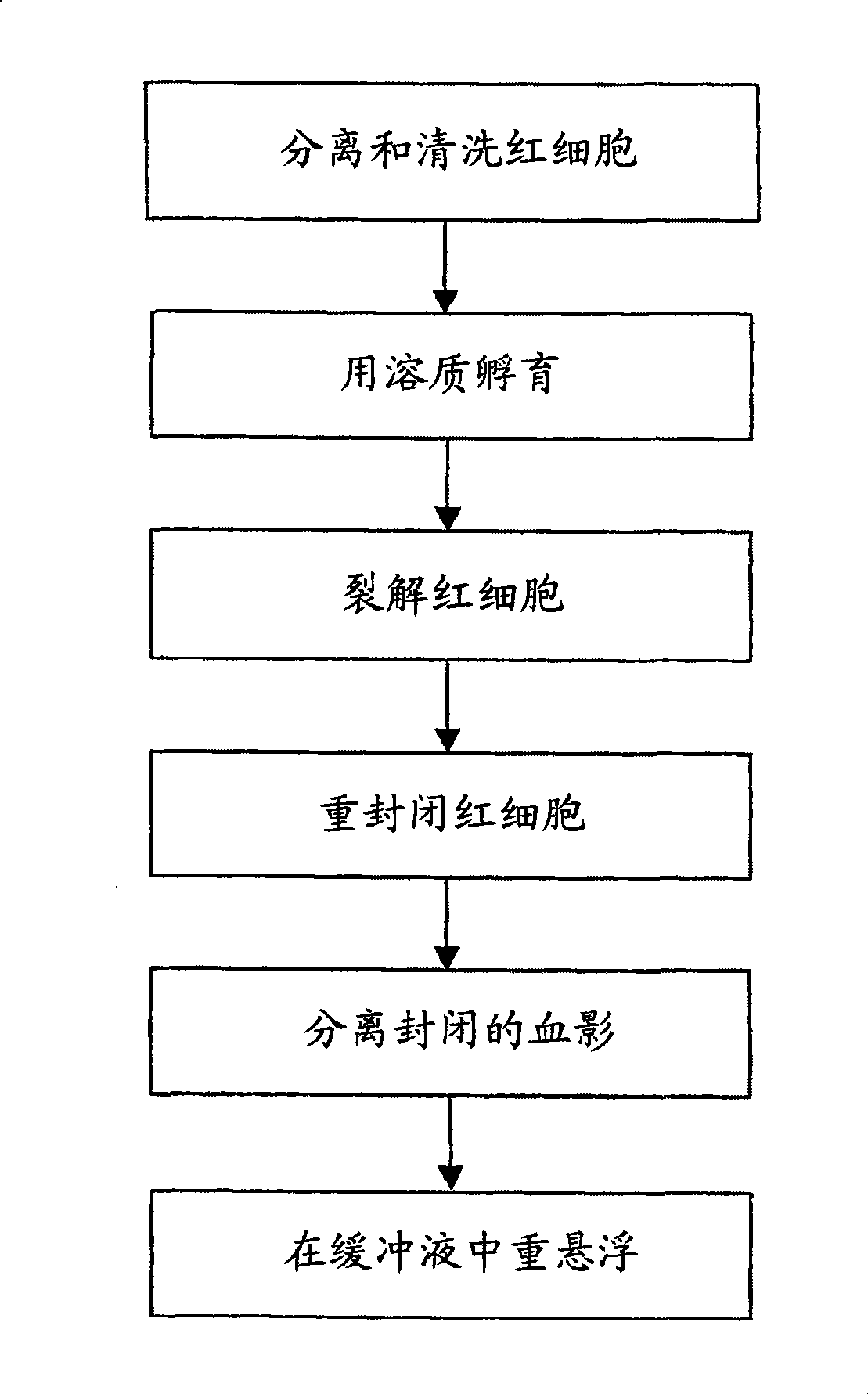
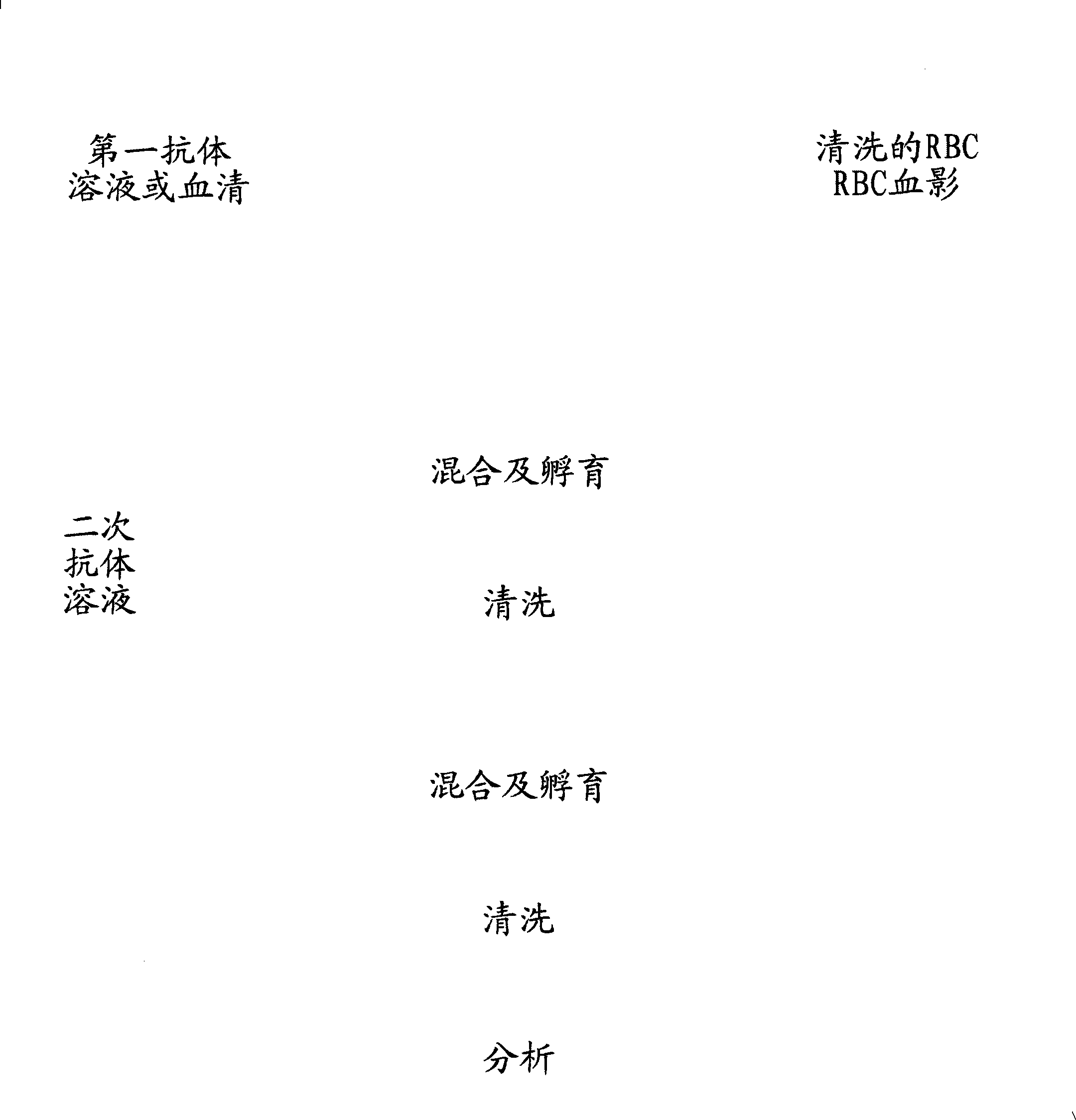
[0016] Example 1 Separation of red blood cells and preparation of labeled ghost cells
[0017] Red blood cell wash-storage buffer
[0018] Sodium chloride = 4.383g
[0019] Monosodium phosphate = 0.345g
[0020] Magnesium chloride hexahydrate = 0.203g
[0021] Benzylsulfonyl chloride=0.0435g, dissolved in 500ml of distilled water.
[0022] Red blood cell lysis buffer:
[0023] Monosodium phosphate = 0.69g
[0024] Magnesium chloride hexahydrate = 0.203g
[0025] Benzylsulfonyl chloride=0.0435g, dissolved in 500ml of distilled water.
[0026] RBC blocking solution:
[0027] Sodium chloride=6g, dissolved in 50ml of distilled water.
[0028] Test plan:
[0029] i) Take 1ml of storage buffer and put it into a 2ml centrifuge tube
[0030] ii) Add 1 drop of fresh blood from a finger prick (~25ul), invert gently to mix
[0031] iii) Centrifuge at ~600-100Og for 2 minutes and remove supernatant. Add 1.5 ml storage buffer, gently resuspend the RBC pellet and repeat centrifu...
Embodiment 2
[0038] Example 2: Generation of libraries of encoded ghost cells
[0039] TAMRA-labeled dextran with a molecular weight of 3000 (labeling density of 1mol / mol) was obtained from Molecular Probes, and FITC-labeled dextran with a molecular weight of 4000 (labeling density of 0.05mol / mol-0.5 / mol / mol) was obtained from Obtained from Sigma-Aldrich Company. Five different stock solutions at concentrations of 10 mg / ml, 2 mg / ml, 0.4 mg / ml, 0.08 mg / ml and 0.016 mg / ml were prepared for each dextran using storage buffer. The recipe in Example 1 was used to prepare 10 different populations of encoded ghost cells. These ghost cells were characterized for their fluorescence using flow cytometry. A 1:1 (v / v) mixture of TAMRA and FITC-dextran was used to generate encoded ghost cells with both encoding dyes, respectively.
Embodiment 3
[0040] Example 3: Generation of Libraries of Encoded Ghost Cells for Magnetic Response
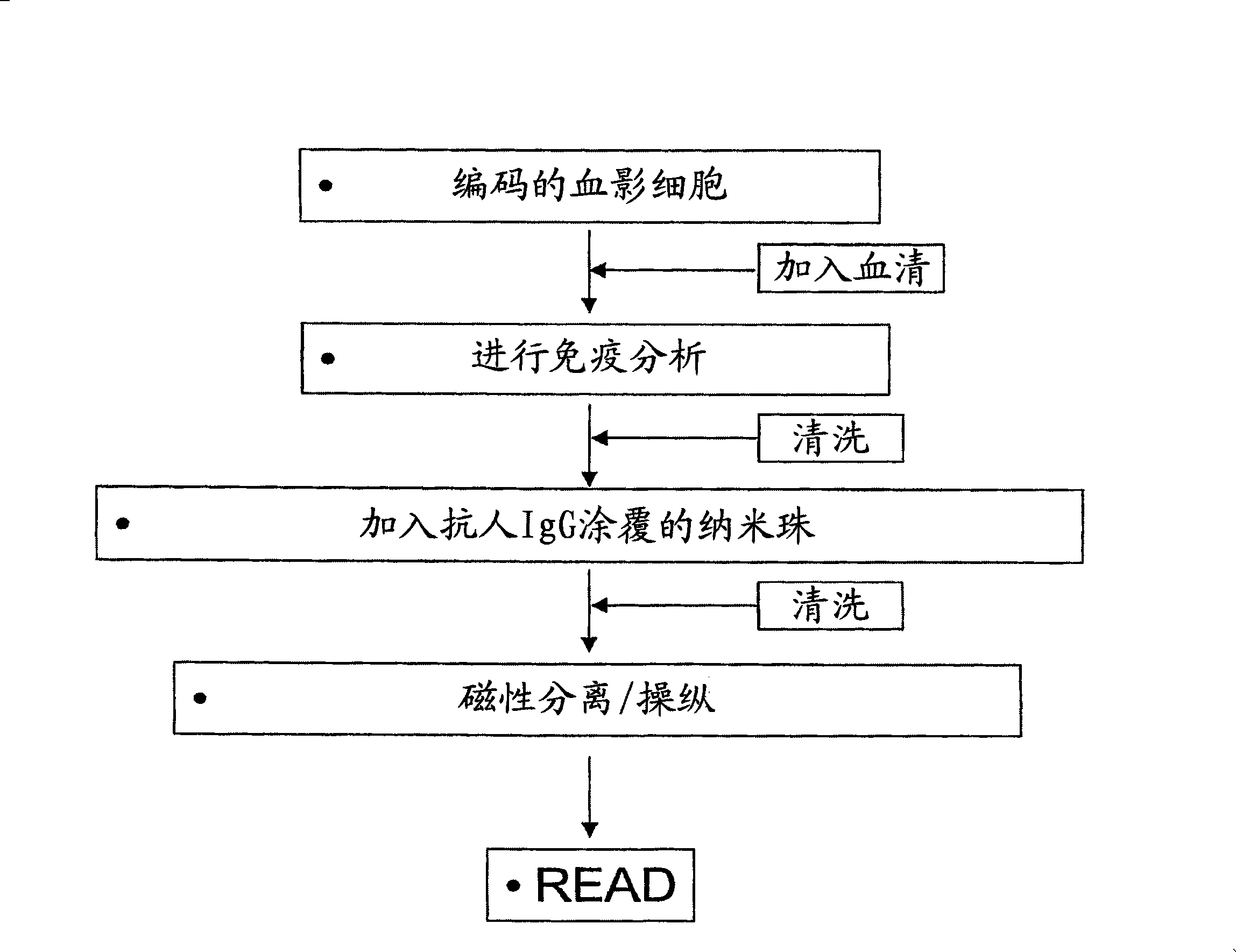
[0041] Encoded ghost cells can be made magnetic by magnetic cell surface labeling methods known in the art. Several companies sell kits for magnetically labeling and isolating target cells (www.miltenyibiotec.com, www.immunicon.com, www.dvnalbiotech.com). Magnetically labeled whole blood cells are also commercially available (www.diagast.com). A particularly contemplated method utilizes anti-human IgG-coated magnetic nanobeads for labeling, which can be added after binding of antibody from the sample, e.g. figure 2 as outlined.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com