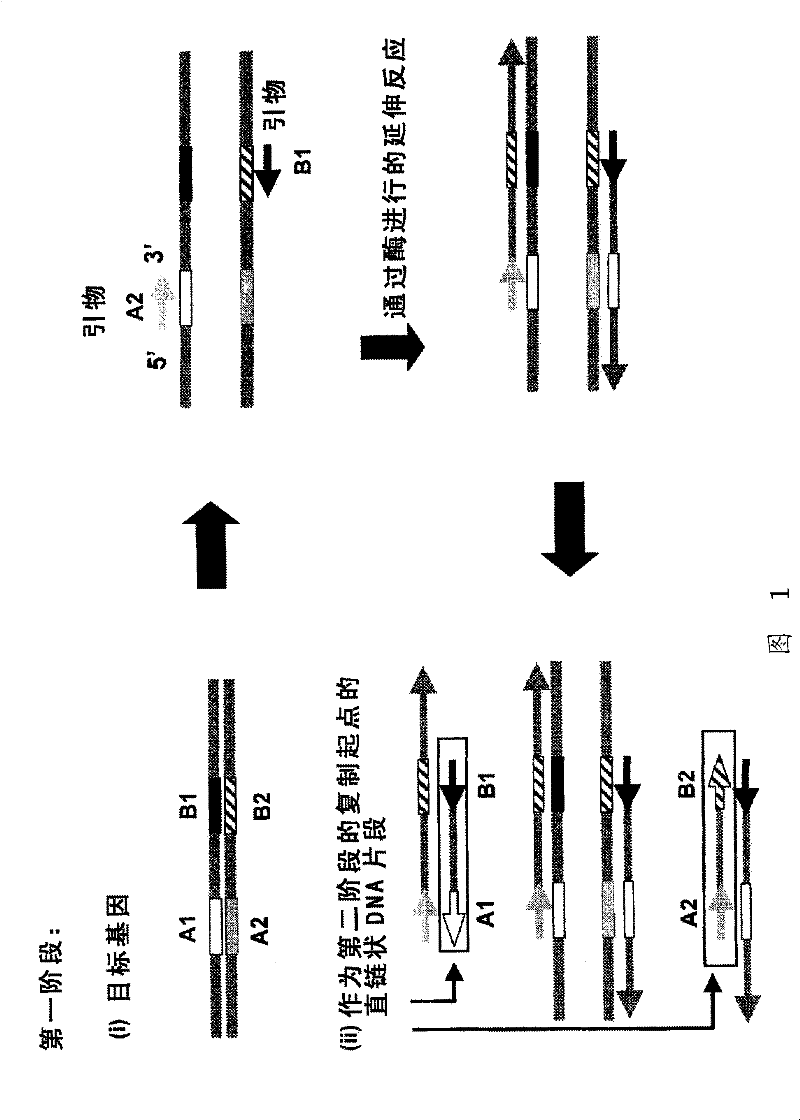
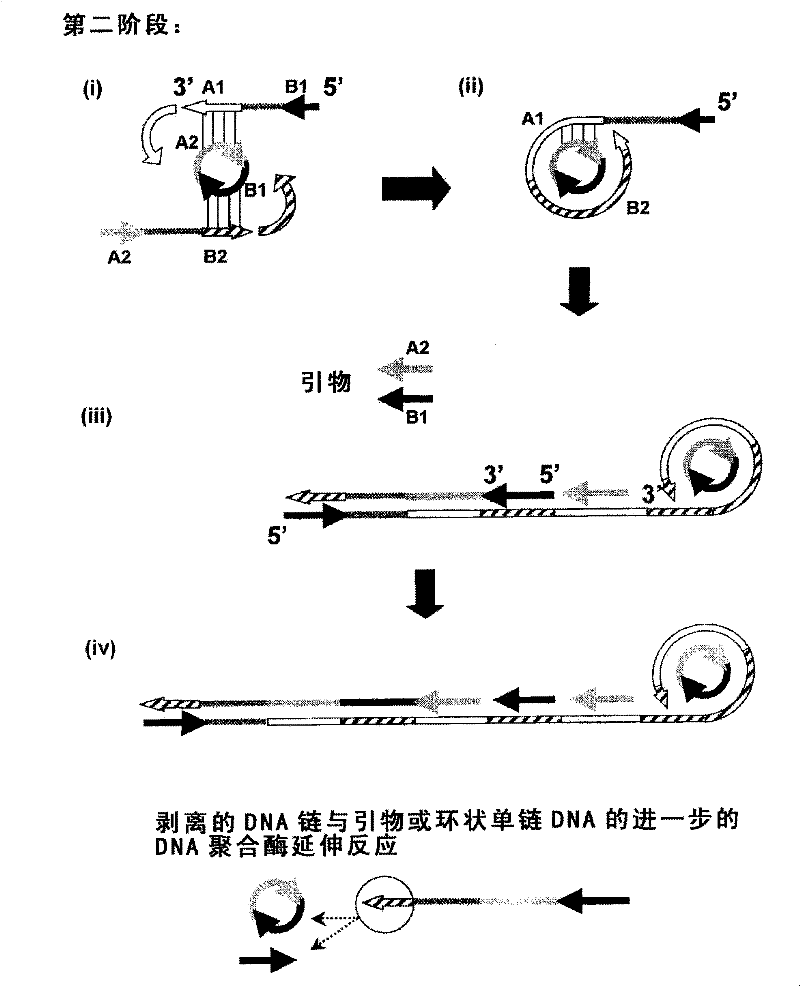
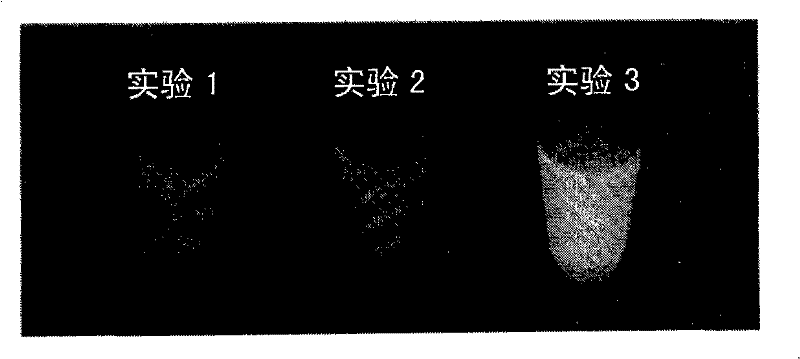
Nucleic acid amplification method
A nucleic acid and nucleic acid molecule technology, applied in the field of nucleic acid amplification, can solve the problems of cost detection method design and operation, detection sensitivity and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0105] In the method for preparing circular single-stranded DNA of the present invention, the "complementary sequence (of the 5'-terminal region and 3'-terminal region)" is preferably the sequence of the 5'-terminal region and the 3'-terminal region of the linear single-stranded DNA. The sequence of the region is completely complementary to the sequence, but there may be a base pair mismatch as long as it can anneal to the aforementioned linear single-stranded DNA. The number of such base pairs depends on the reaction conditions and the length of the entire region. For example, it may be 1, 2, or 3 in each region, or may be about 1% to 50% of the total bases in each region. However, those skilled in the art can understand that, in order to perform ligation efficiently, it is preferable that the bases at the 5' end and the 3' end that are actually subjected to the ligation reaction by ligase each form a base with the base of the adapter during circularization. base pair.
[01...
Embodiment
[0124] Substances without a company name after the drug name are manufactured by Wako Pure Chemical Industries, Ltd. In addition, unless otherwise specified, the solvent is water.
[0125] (TE buffer composition)
[0126] 4mmol / l Tris-HCl (trishydroxymethylaminomethane) hydrochloric acid
[0127] 1mmol / l disodium ethylenediaminetetraacetic acid dihydrate (EDTA)
[0128] Adjust to pH8.0
[0129] (1.2% TAE gel for electrophoresis)
[0130] 1.2% agarose
[0131] 4mmol / l Tris-HCl
[0132] 1mmol / l EDTA
[0133] 1mmol / l acetic acid
[0134] (2.0% TAE gel for electrophoresis)
[0135] 2.0% agarose
[0136] 4mmol / l Tris-HCl
[0137] 1mmol / l EDTA
[0138] 1mmol / l acetic acid
example 1
[0140] [Preparation method of target]
[0141] Enzyme preparations and kit preparations shown below were in accordance with the instructions for use unless otherwise specified.
[0142] After treating pUC19 (Takara Bio Inc.) with the restriction enzyme DraI (Takara Bio Inc.), electrophoresis was performed using a 1.2% TAE gel for electrophoresis (100 V, 50 minutes) using an electrophoresis tank Mupid-ex (Advance).
[0143] Then, stain with nucleic acid staining reagent SYBR GreenI (manufactured by CAMBREX company), cut out the target DNA fragment (about 2kb) from the gel, and recover it with the DNA extraction kit NucleoS pin Extract kit (MACHEREY-NAGEL company) to form the target preparation solution (10 mmol / l).
[0144] [Synthesis of circular single-stranded DNA]
[0145] The primers used for circular single-stranded DNA synthesis are listed in Table 1. 43C is a primer for phosphorylation modification at the 5' end, and Bind is a primer for cartridge purification (manufac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com