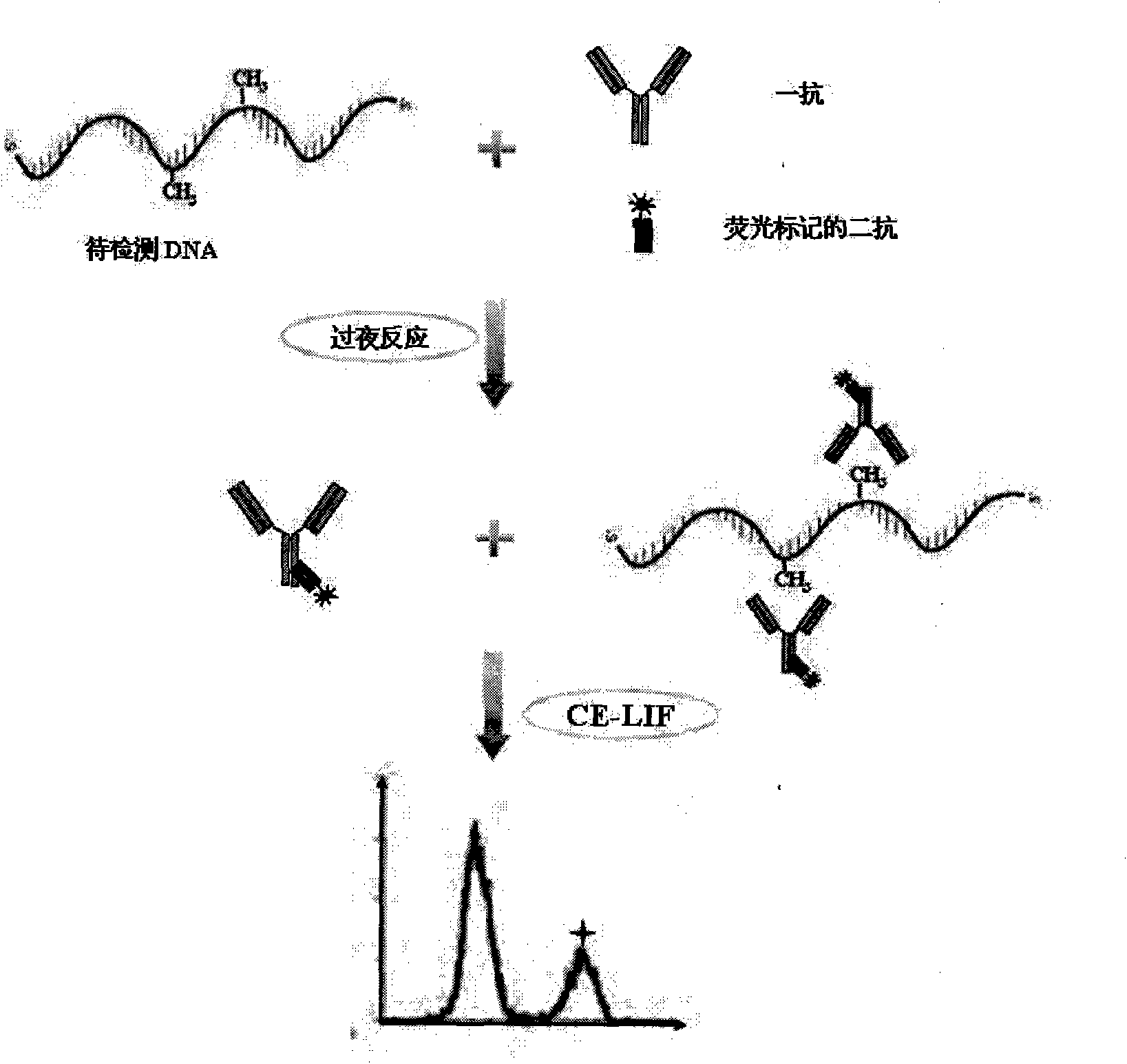
Integral genome DNA methylation level analysis based on immune capillary electrophoresis-laser induced fluorescence
A laser-induced fluorescence and capillary electrophoresis technology, which is applied in the field of biological detection, can solve the problems of complicated DNA enzymatic hydrolysis operation and limited analysis, and achieves the effect of overcoming the complicated DNA operation, the small amount of DNA, and the wide application prospect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
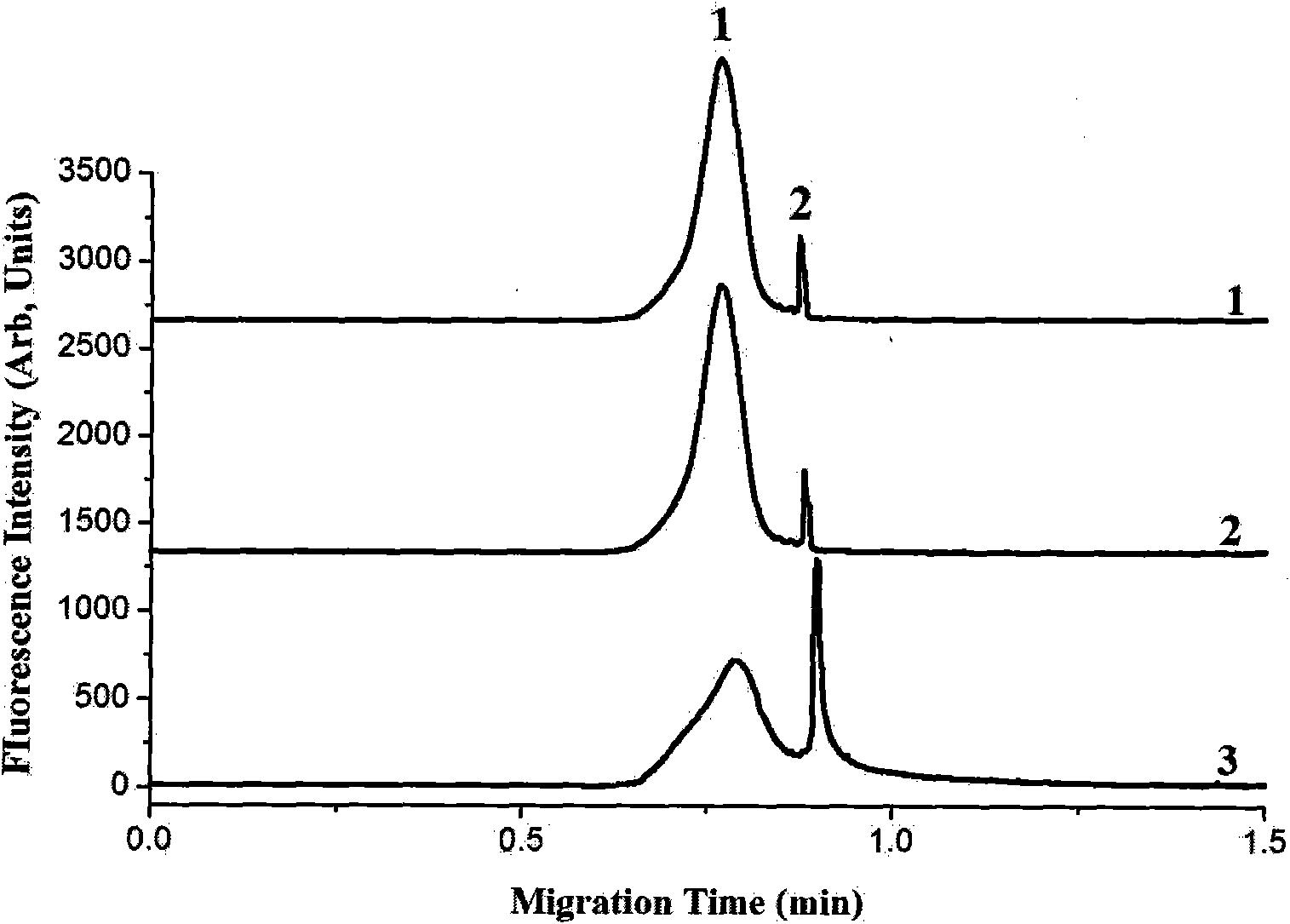
[0032] Example 1. Immuno-CE-LIF detection of overall methylation level in lambda DNA
[0033] In this example, the primary antibody is a mouse-derived 5-methylcytosine monoclonal antibody, which can specifically recognize and bind to 5-methylcytosine (5mC) in single-stranded DNA; Alexa Fluor 546 fluorescently labeled Fc is specific The F(ab') fragment of anti-IgG1 is the secondary antibody used in this example. After the λDNA to be tested was treated at 95°C for 5 minutes, the double-stranded DNA was denatured into a single strand, and placed on ice for 10 minutes to prevent the renaturation of the single-stranded DNA, and reacted with the above-mentioned primary antibody and secondary antibody overnight at 4°C to form a DNA-antibody immune complex. The reaction system of DNA and antibody was 20 μL, and the concentrations of primary antibody and secondary antibody were 1 μg / mL and 2 μg / mL, respectively.
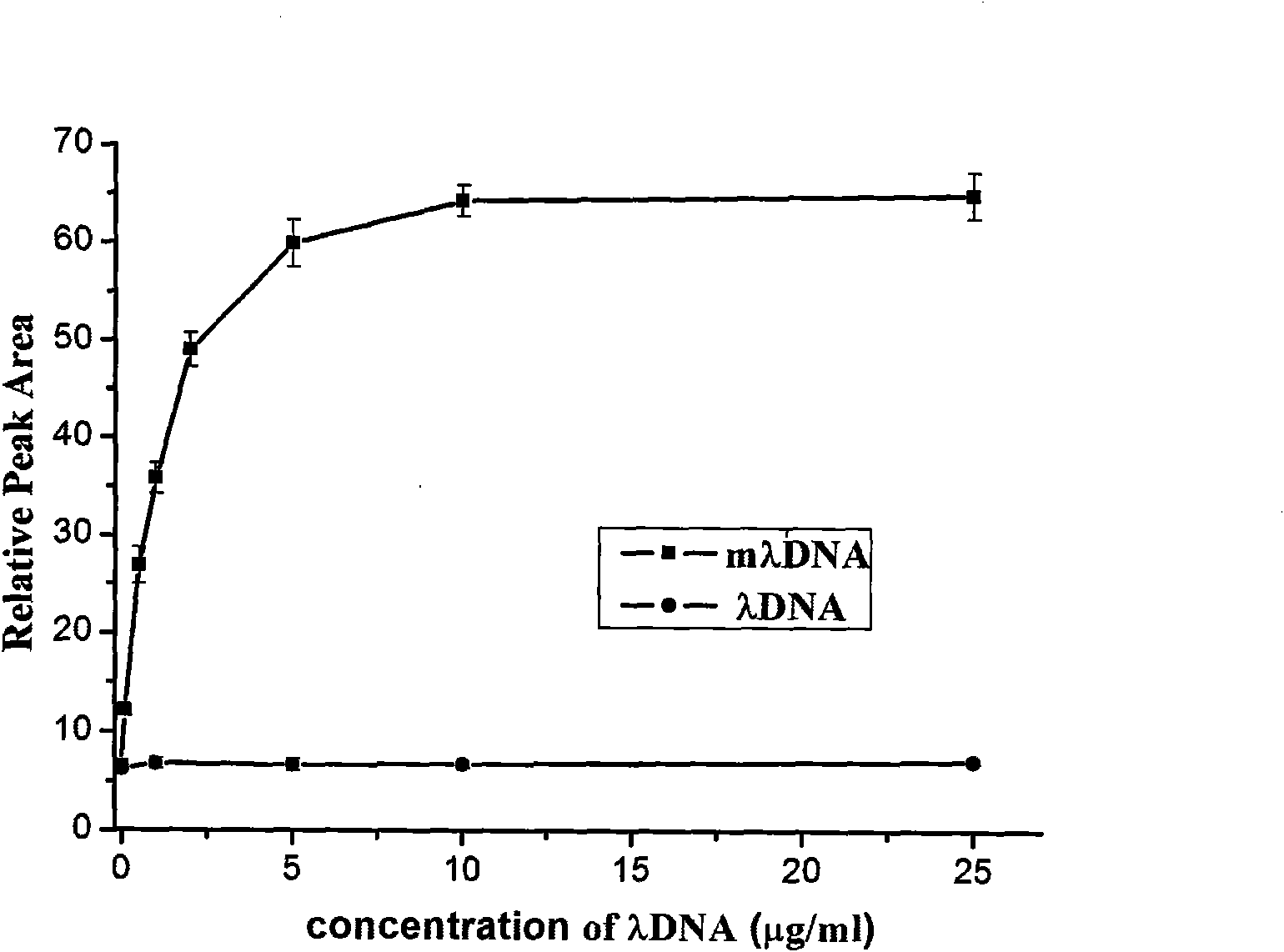
[0034] The reaction mixture was separated in a CE-LIF device with an ex...
Embodiment 2、A549
[0039] Example 2. Immuno-CE-LIF detection of the overall methylation level of genomic DNA in A549 cells
[0040] 50 μg / mL of A549 cell genomic DNA was denatured into single-stranded DNA, reacted with 5-methylcytosine monoclonal antibody and fluorescently labeled secondary antibody overnight at 4°C, and carried out immuno-CE-LIF analysis according to the conditions described in Example 1. attached Figure 5 For the detection of the overall methylation level of A549 cells, the electrophoresis peak type is similar to the peak type of mλDNA, indicating that the genomic DNA of A549 cells can react with antibodies to form obvious DNA-antibody immune complexes. attached Figure 6 It shows that when the concentration of genomic DNA in A549 cells is in the range of 0-50 μg / mL, CE-LIFP detection is performed, and the amount of DNA-antibody immune complex has a good linear relationship with the DNA concentration (R 2 =0.99), the example shows that the present invention can be used to d...
Embodiment 3
[0041] Example 3, CE-LIF detection of the effect of 5-Aza-dC on the overall methylation level of cellular genomic DNA
[0042] After exposing A549 cells and HepG2 cells to different concentrations of 5-Aza-dC for 72 hours, genomic DNA was extracted respectively, and 50 μg / mL of cellular genomic DNA was reacted with antibodies overnight, and immuno-CE-LIF detection was carried out according to the above method. attached Figure 7 It shows the effect of different concentrations of 5-Aza-dC on the overall methylation level of genomic DNA in A549 cells and HepG2 cells. As the concentration of 5-Aza-dC increases, the overall DNA methylation level of cells shows a downward trend. At the highest exposure concentration (10 μM), the overall methylation level of genomic DNA in A549 cells and HepG2 cells decreased by 64% and 46%, respectively, and when the exposure concentration of 5-Aza-dC was 0.01 μM, the overall methylation level of DNA was inhibited ( 10%-20%) can be obtained throug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com