Oligonucleotide aptamer group for specifically identifying staphylococcus aureus and use thereof
A staphylococcus and oligonucleotide technology, applied in the application field of identifying staphylococcus aureus, can solve difficult and time-consuming problems, achieve the effects of low preparation cost, simple operation, and improved sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
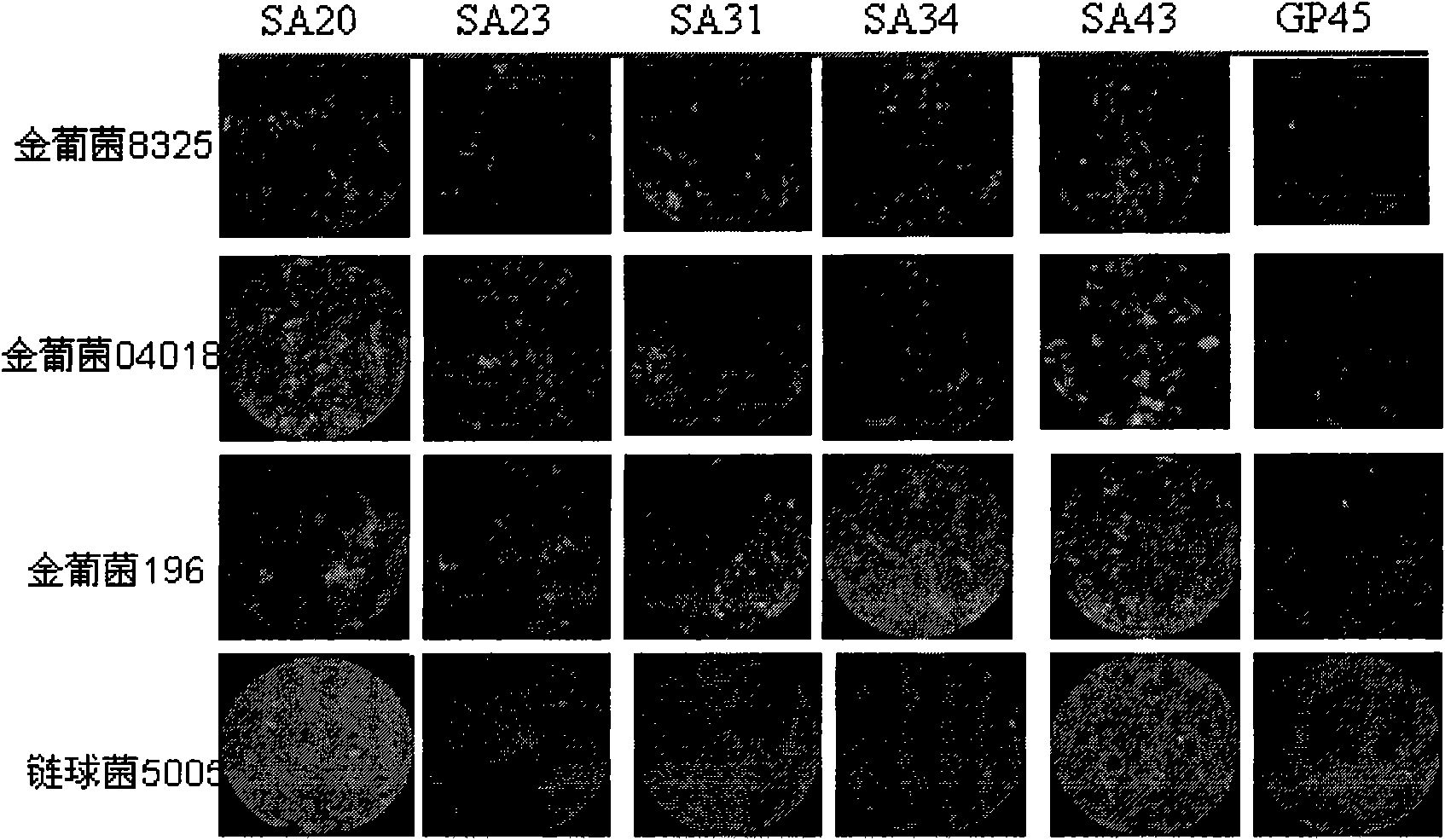
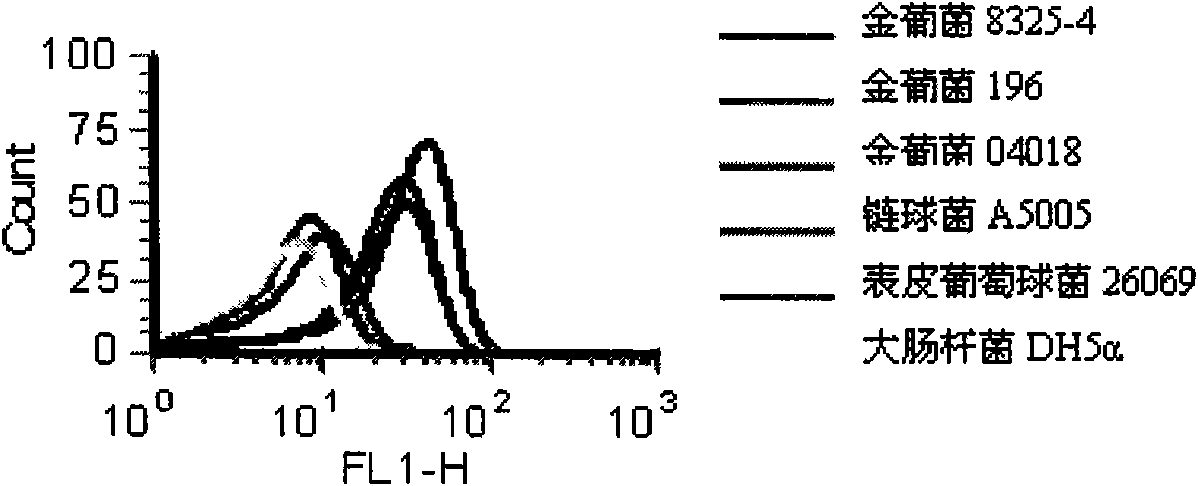
[0016] The present invention will be further described below by subtractive SELEX screening to obtain Staphylococcus aureus-specific oligonucleotide aptamers and comparing the recognition and detection rates of five specific oligonucleotide aptamers to Staphylococcus aureus.
[0017] 1. Synthesize a random single-stranded DNA library and primers (completed by Invitrogen).
[0018] Library GP45: 5'-GCAATGGTACGGTACTTCC(45N)CAAAAGTGCACGCTACTTTGCTAA-3'
[0019] 5' Primer (Plong-1): 5'-GCAATGGTACGGTACTTCC-3'
[0020] 3' Primer (P11): 5'-TTAGCAAAGTAGCGTGCACTTTTG-3'
[0021] 3' Primer (Pstem-loop3): 5'-GCTAAGCGGGTGGGACTTCCTAGTCCCACCCGCTTAGCAAAGTAGCGTGCACTTTTG-3'
[0022] 2. Acquisition and processing of bacterial targets for screening.
[0023] Staphylococcus aureus, Streptococcus and Staphylococcus epidermidis were cultured in TSB medium, and they were shaken at 37°C until early logarithmic growth (OD 600 about 0.2). Then collect the bacteria separately, take 100μl bacterial so...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com