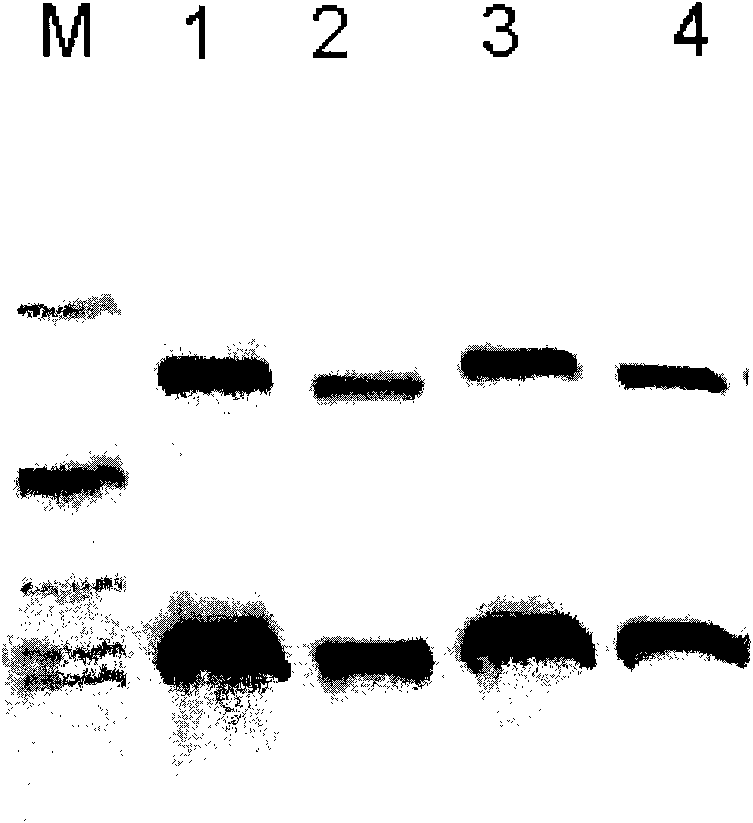Recombinant fusion proteins containing p53 genes, recombinant and application thereof
A technology of p53 gene and fusion protein, applied in medical preparations containing active ingredients, drug combinations, peptide/protein components, etc., can solve the problems of tumor cells being invasive and prone to distant metastasis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0068] Example 1, preparation of p53 gene
[0069] Take 1g of human fetal total RNA, and add the reaction components sequentially according to the operation guide of cDNA synthesis system. The total reaction volume is 20 μl, reacted at 42°C for 15 minutes, then inactivated at 95°C for 5 minutes, and used as a PCR template. Using P7: GAGAATTCATGGAGGAGCCGCAGTC; P4: CCGTCGACTTAGTCTGAGTCAGGC as primers, PCR human full-length p53 gene.
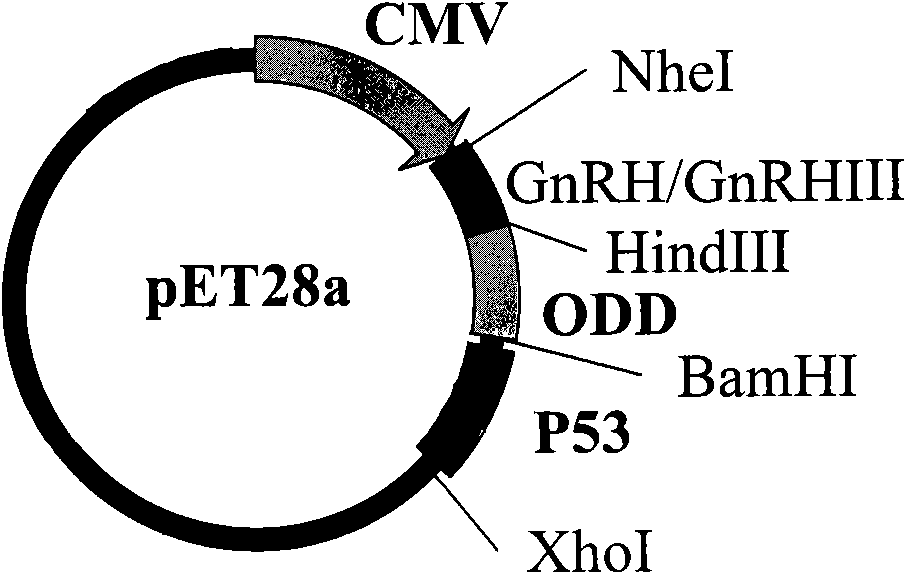
[0070] Take the PCR fragments of vector pET28a and p53 respectively, carry out double digestion with EcoR I and SalI, treat at 37°C for 3h, and then recover with gel recovery kit. The pET carrier fragment and p53 fragment were mixed at a ratio of 1:3, added with 3 U of T4 ligase, reacted at 16°C for 16 hours, and transformed the reaction product into competent Escherichia coli DH5α, picked the successfully transformed colony, and extracted the plasmid for PCR identification. The positive clone was named pET-p53 after correct sequencing. The pE...
example 2
[0071] Example 2, preparation of GnRH-ODD-p53 gene and GnRHIII-ODD-p53 gene
[0072]GnRH and GnRHIII genes were constructed by overlap extension method. Equal amounts of GP1: CTAGCATGGAGCACTGGTCCTATGGACTGCGCCCTGGAA and GP2: AGCTTTCCAGGGCGCAGTCCATAGGACCAGTGCTCCATG were mixed into the PCR reaction system and reacted 30 times to obtain the fragment (GnRH).
[0073] In the same way, G3P1:
[0074] CTAGCATGGAGCACTGGTCCCACGACTGGAAGCCTGGAA and G3P2:
[0075] An equal amount of AGCTTTCCAGGCTTCCAGTCGTGGGACCAGTGCTCCATG was mixed and added into the PCR reaction system, and reacted 30 times to obtain the fragment (GnRHIII).
[0076] Mix the pET-p53 vector digested with Nhe I and HindIII with GnRH or GnRHIII fragments at a ratio of 1:3, add T4 ligase, react at 16°C for 16 hours, transform the reaction product into competent Escherichia coli DH5a, pick Successfully transformed colonies were identified by plasmid PCR. The positive clones were correctly named pET-GnRH-p53(GP) and pET-Gn...
example 3
[0078] Example 3, preparation of GnRH-ODD-p53 and GnRHIII-ODD-p53 fusion protein
[0079] 1), expression of GnRH-ODD-p53 and GnRHIII-ODD-p53
[0080] Pick the positive colonies transformed into pET-GnRH-p53 and pET-GnRHIII-p53 respectively and place them in LB medium (containing kanamycin 60g / ml) for activation, then inoculate them in 2YT medium according to 5-20%, and cultivate until OD 600 When the value is 0.5-2.0, add IPTG to 0.4-1mM to induce expression. Collect the bacterial cells induced to express for 3-24 hours, and wash with PBS.
[0081] 2), Western blot identification of GnRH-ODD-p53 and GnRHIII-ODD-p53
[0082] The fusion protein obtained by induced expression was subjected to polyacrylamide gel electrophoresis (12% SDS-PAGE, see image 3 ) analysis showed that there was target protein in the precipitate, and its molecular weight was consistent with the theoretical value. After western blotting, it was identified and analyzed as GnRH-p53 series fusion p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com