Manufacturing method of adjustable liver damage animal model and special DNA fragment thereof
A technology of DNA molecules and fragments, which is applied in the production of adjustable liver injury animal models and the field of special DNA fragments, which can solve the problems of slow speed, incomplete induction, and difficult operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1, the construction of pAlb-rtTA-polyA plasmid
[0069] 1. Construction of Psk-rtTA-polyA plasmid
[0070] 1. Digest the pTet-On-Advanced vector (Clontech Company, Catalog No.: 630930) with restriction endonucleases EcorI and HindIII at 37°C overnight. After the digestion, the digested product is subjected to 1% agarose gel electrophoresis. The rtTA-polyA fragment of about 1.2kb was recovered with Dingguo glue recovery kit. The recovered DNA fragments were dissolved in 10ul ultrapure water. The recovered DNA fragments were sequenced, see sequence 1 in the sequence listing, and the amino acid sequence encoded by the rtTA gene was shown in sequence 2 in the sequence listing.
[0071] 2. Use restriction enzymes EcorI and HindIII to digest the pBluscript vector (Fermentas company) overnight at 37°C. After the digestion, the digested product is subjected to 1% agarose gel electrophoresis, and about 3kb is recovered with the Dingguo gel recovery kit pSK vector b...
Embodiment 2
[0085] Embodiment 2, making TRE-PminCMV-uPA adenovirus
[0086] 1. Amplification of the prourokinase activator (uPA) gene
[0087] A pair of primers were designed according to the mouse uPA mRNA sequence (NM_008873) in Gene bank as follows:
[0088] Upstream primer (29bp): 5'-GGC GCTA ATGAAAGTCTGGCTGGCGAG-3';
[0089] Downstream primer (29bp): 5'-TCT GGTACC GAGAGGACGGTCAGCATGGG-3'.
[0090] Among the above primers, the underlined ones are restriction sites, and the italic boldfaced ones are Kozak sequences for enhanced expression. The upstream primer is located at the initiation ATG of uPA mRNA and contains an NheI restriction site. The downstream primer is located 76bp downstream of the stop codon and contains a Kpn I restriction site.
[0091]1. Extract kidney RNA of CD-1 mouse (purchased from Weitong Lihua) and reverse transcribe into cDNA. Using cDNA as a template, using the above primers, use TaKaRa's Pyrobest DNA Polymerase to perform PCR reaction.
[0092] 2....
Embodiment 3
[0142] Embodiment 3, preparation of liver injury mice
[0143] 1. Preparation of Alb-rtTA mice
[0144] (1) Digestion, recovery and purification of Alb-rtTA fragments
[0145] 1. Digest pAlb-rtTA-polyA plasmid overnight at 37°C with restriction enzymes SacI and KpnI.
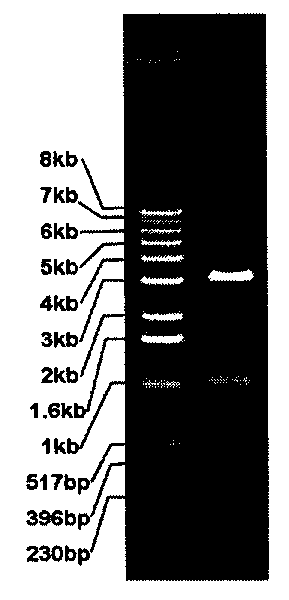
[0146] 2. The digested products were subjected to 1% agarose gel electrophoresis. Electrophoresis results see Figure 5 . The 3.5kb band was cut out, and the DNA was recovered with a DNA gel recovery kit (Dingguo).
[0147] 3. Use Promega Wizard DNA clear-up kit (Cat: A 7280) to further purify the DNA fragment.
[0148] 4. TE (7mM Tris-HCl, 0.15mM EDTA, pH 7.5) dissolves the purified DNA.
[0149] (2) Preparation of DNA samples for pronuclear injection
[0150] To measure the OD value of the DNA sample, dilute the DNA to 2-5ng / ul with TE; centrifuge at 12000g×10min (4°C); carefully suck out the upper 1 / 3 part of the solution after centrifugation, divide it into several centrifuge tubes, freeze Store in -...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com