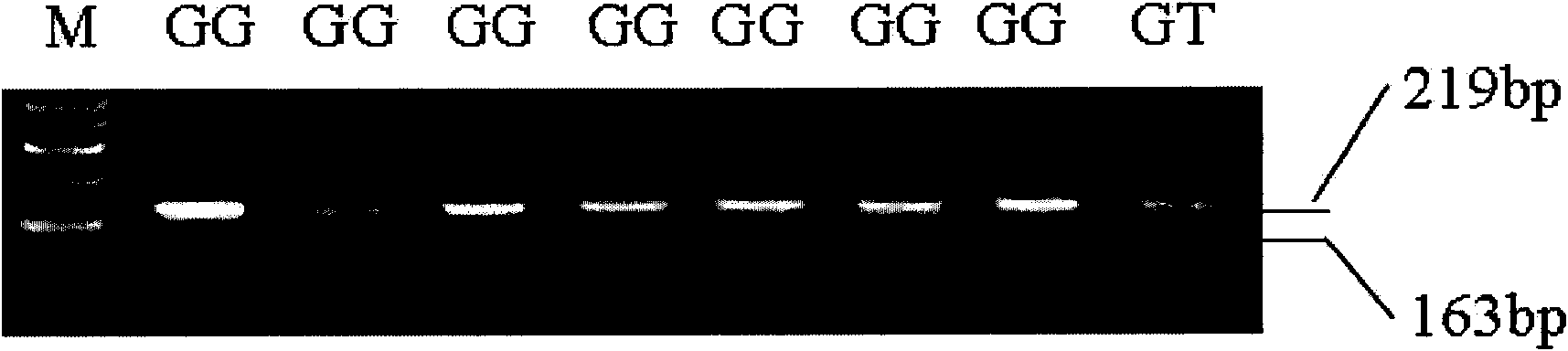Sheep back fat trait-related SNP and application thereof
A technology of sheep and chain reaction, applied in the field of bioengineering, can solve problems such as unclear knowledge, marker-assisted selection and breeding obstacles of sheep, and less research on CAST genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] (1) Fully cut up about 0.3 g of muscle tissue with ophthalmic scissors, put it in a 1.5 ml centrifuge tube, and dry it for 20 minutes.
[0019] (2) Add 500 μl of tissue DNA extraction solution to the above centrifuge tube.
[0020] (3) Add RNase solution to the above centrifuge tube to a final concentration of 20 μg / ml, mix thoroughly, and digest at 37°C for 1-2 hours.
[0021] (4) Add proteinase K to the above centrifuge tube to a final concentration of 150 μg / ml, mix thoroughly, and digest at 55°C for 12-24 hours.
[0022] (5) Add an equal volume of Tris saturated phenol to the above centrifuge tube, slowly invert the centrifuge tube for 10 minutes to fully mix the two phases of the solution at 4°C, centrifuge at 12000rpm for 10 minutes, then transfer the supernatant to a clean centrifuge tube . repeat.
[0023] (6) Add an equal volume of Tris-saturated phenol:chloroform:isoamyl alcohol (25:24:1), mix well, and centrifuge at 12,000 rpm for 10 minutes at 4°C.
[00...
Embodiment 2
[0030] The PCR primers for PCR-SSCP are shown in the table below.
[0031] 1. PCR amplification
[0032] Table 1 5 pairs of specific primer sequences, annealing temperature and target fragment length
[0033]
[0034] Add to PCR tube:
[0035] Template cDNA 1.0μl
[0036] Primer 1 (20pmol / μl) 0.3μl
[0037] Primer 2 (20pmol / μl) 0.3μl
[0038] 10mM dNTP Mix 2.0μl
[0039] 10× PCR buffer 2.5 μl
[0040] Taq DNA polymerase 0.30μl
[0041] wxya 2 O make up to 25 μl
[0042] PCR amplification temperature setting: pre-denaturation at 95°C for 3min
[0043] 35 cycles: denaturation at 95°C for 25 sec; annealing temperature see the above table, annealing for 25 sec; extension at 72°C for 20 sec;
[0044] Extend at 72°C for 10min
[0045] Cool down to 4°C for storage
[0046] 2% agarose gel was used to detect the results by electrophoresis. The results showed that the PCR products of the designed 3 pairs of primers were specific amplification, the fragment length was con...
Embodiment 3
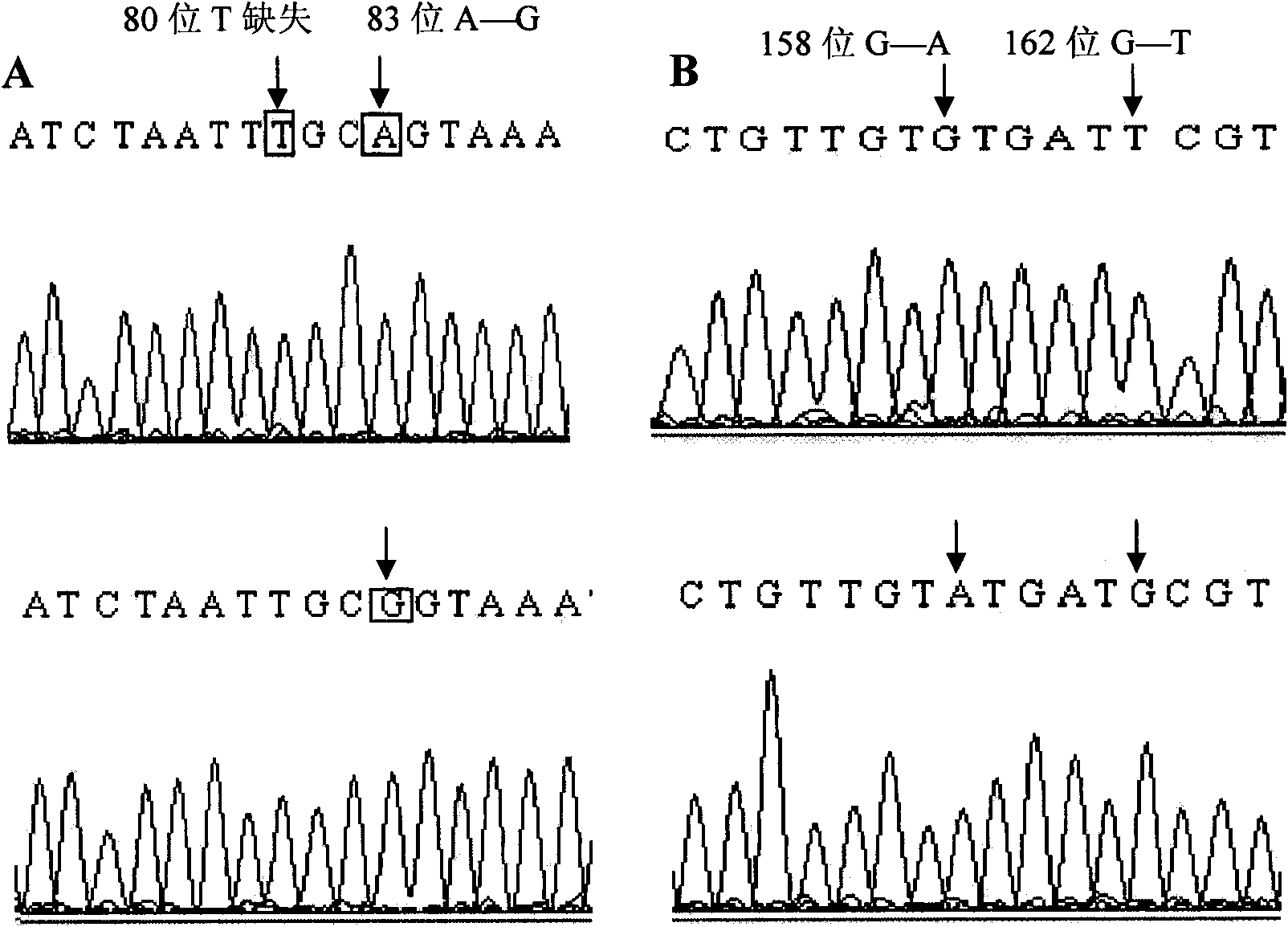
[0065] According to the method described in Example 2, PCR amplification was performed on the total DNA of 450 offspring of Dorper sheep×Hu sheep, 145 offspring of Dorset sheep×Hu sheep, and 45 offspring of Suffolk sheep×Xinjiang fine-wool sheep. For the CAST-S3 amplification product, the HinfI enzyme can recognize the mutation at position 162, and the PCR product is digested with HinfI enzyme and typed ( image 3 ).
[0066] The digestion system is 20 μl, which contains 10 μl of PCR product, 2 μl of 10× restriction endonuclease-specific buffer, 4 U of restriction endonuclease, and high-pressure sterilized double-distilled water to make up to 20 μl. After mixing, digest at 37°C for about 4 hours according to the instructions of the enzyme. The digested products were electrophoresed on 2.0% agarose gel at 5V / cm, observed under ultraviolet light, and photographed.
[0067] The linear model software in SASv8.02 (Statistical Analysis System) was used to analyze the correlation b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com