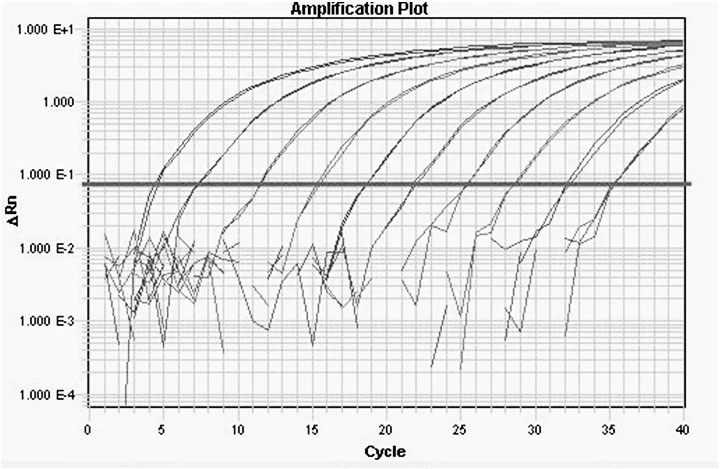
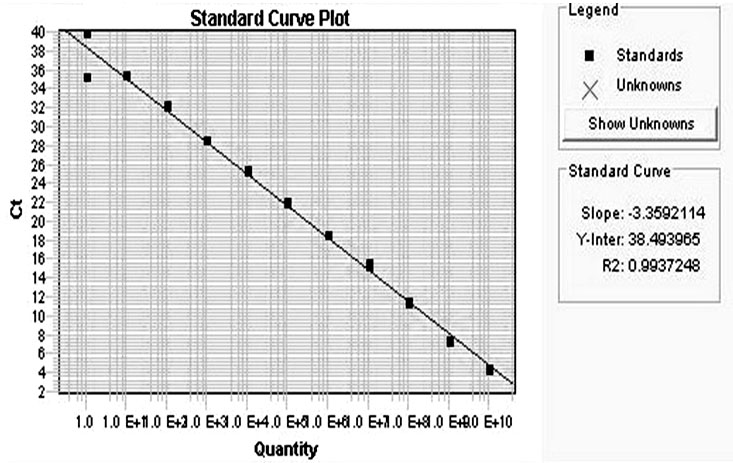
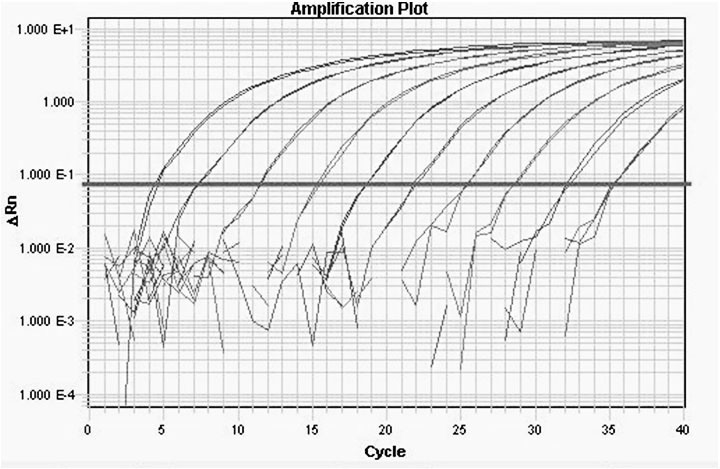
Fluorescent quantitative RT-PCR detection method of M.tuberculosis-complex
A technique for Mycobacterium tuberculosis and fluorescence quantification, which is applied in the determination/inspection of microorganisms, fluorescence/phosphorescence, biochemical equipment and methods, etc. Reliable source, avoid use, avoid contamination effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Designing primers and Taqman probes
[0031] Experimental steps:
[0032] In the RDP (Ribosomal Database Project) ribosomal sequence analysis database, the complete 16srRNA sequence of the Mycobacterium tuberculosis complex was searched, and a conserved segment of the sequence was selected through bioinformatics comparison analysis, and a primer express software and primer premier 5.0 software were used to design a Reverse transcription primers, a pair of PCR primers and a Taqman probe. The fluorescent reporter group at the 5' end of the probe is FAM, and the fluorescent quencher group at the 3' end is TAMRA. The primers and probe are located in the Mycobacterium tuberculosis complex Group 16srRNA region, the target amplification fragment is 100 bp.
[0033] The primer and probe sequences are:
[0034] Reverse transcription primer: CCGTATCTCAGTCCCAGTGT
[0035] Upstream primer: MTBC-F: GGGATGCATGTCTTGTGGTG
[0036] Downstream primer: MTBC-R: CCGTCGTCGCCTTGG...
Embodiment 2
[0038] Example 2: Construction and preparation of quantitative standards
[0039] Implementation steps
[0040] 1. Construction of quantitative standards
[0041] The Mycobacterium tuberculosis complex (MTBC) gene fragment containing the target sequence was cloned by RT-PCR, recombined into the plasmid vector pGEM-T Easy, and the DNA sequence was determined. The constructed recombinant plasmid was used as a quantitative standard for detecting the Mycobacterium tuberculosis complex and was named pGEM-T Easy-16srRNA.
[0042] 2. Preparation of Quantitative Standards
[0043] After the plasmid pGEM-T Easy-16srRNA was extracted and purified, the concentration of the plasmid was calculated according to the OD260, OD280 and OD230 values measured by the ultraviolet spectrophotometer, the purity of the plasmid was detected, and the copy number was converted according to the Avogadro constant.
[0044] Calculated as follows:
[0045] Plasmid concentration (ug / ul) = OD260 X diluti...
Embodiment 3
[0050] Example 3: Isolation and extraction of bacteria in sputum samples
[0051] 1. Pretreatment of Sputum Samples
[0052] (1) Take the morning sputum and place it in a sterile sealed bottle, and treat it with NaOH.
[0053] (2) Add an equal volume of 10% NaOH solution, vortex and oscillate to mix well, and put it in a 37°C water bath for digestion for more than 30 minutes until the sputum is completely digested;
[0054] (3) Mix the phlegm and digestive fluid, take 1 ml of the liquefied phlegm sample, centrifuge at 12000 rpm for 5 min in a clean 1.5ml EP tube, and remove the supernatant;
[0055] (4) Wash the precipitate from the previous step with 1ml of cleaning solution (10mM EDTA or normal saline), vortex and mix well, centrifuge at 12000 rpm for 5min, remove the supernatant and keep the precipitate for inspection.
[0056] 2. Simultaneous extraction of RNA and DNA in bacteria using a new extraction method
[0057] (1) Bacterial lysis: Resuspend the precipitate obtai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com