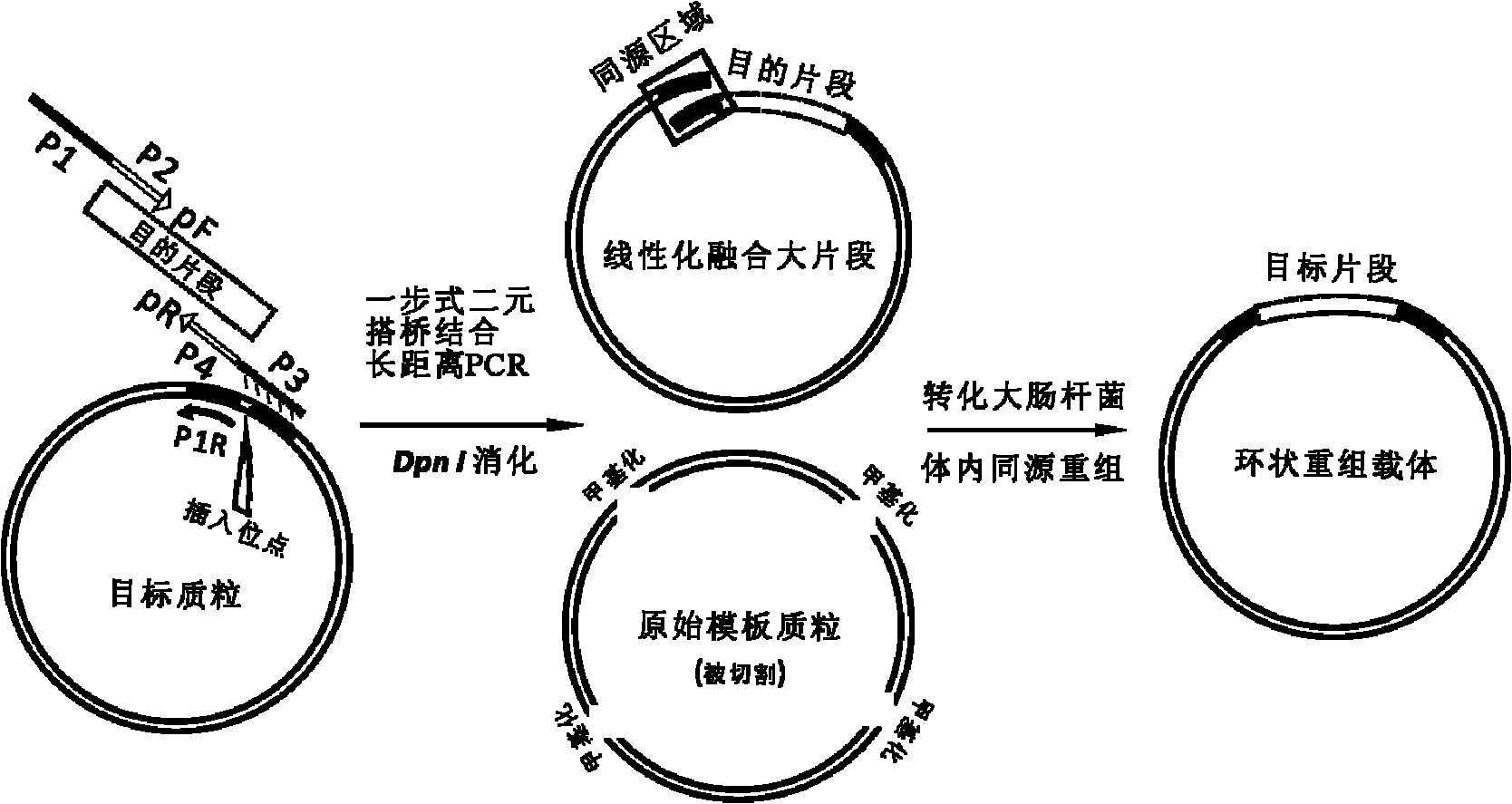
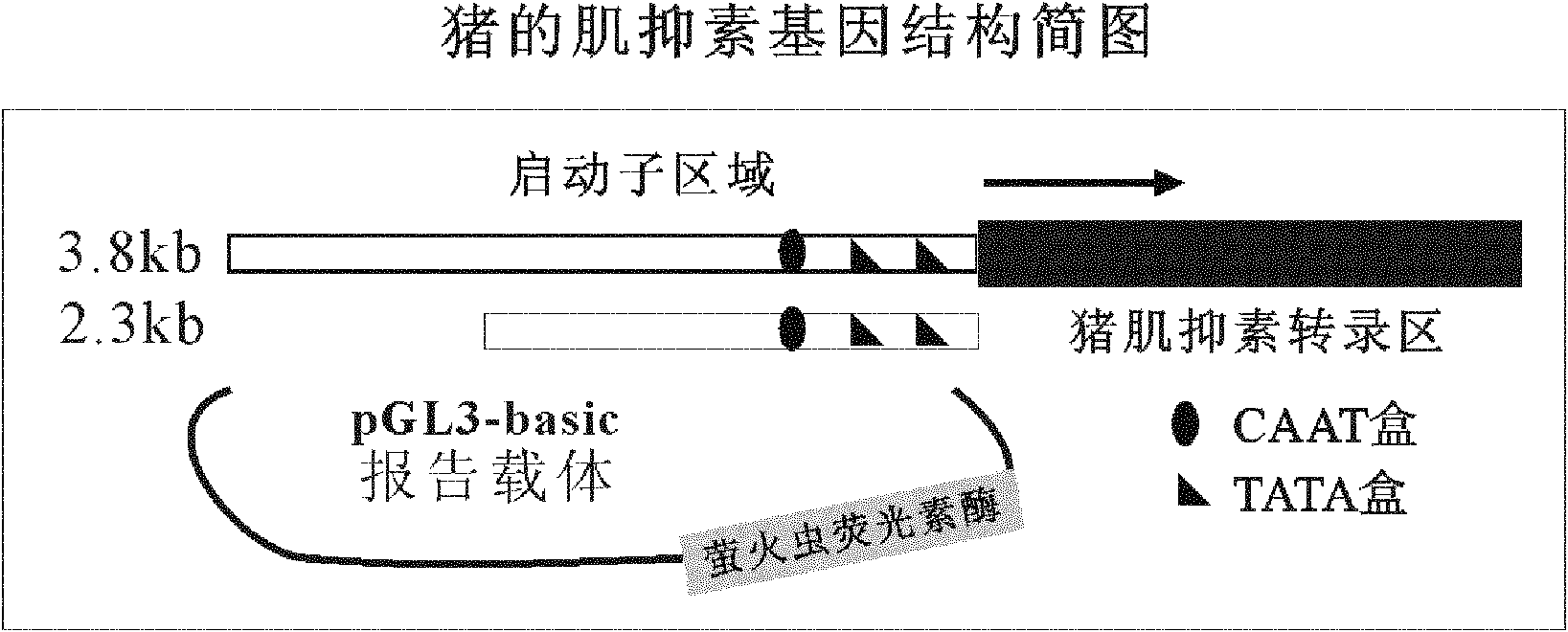
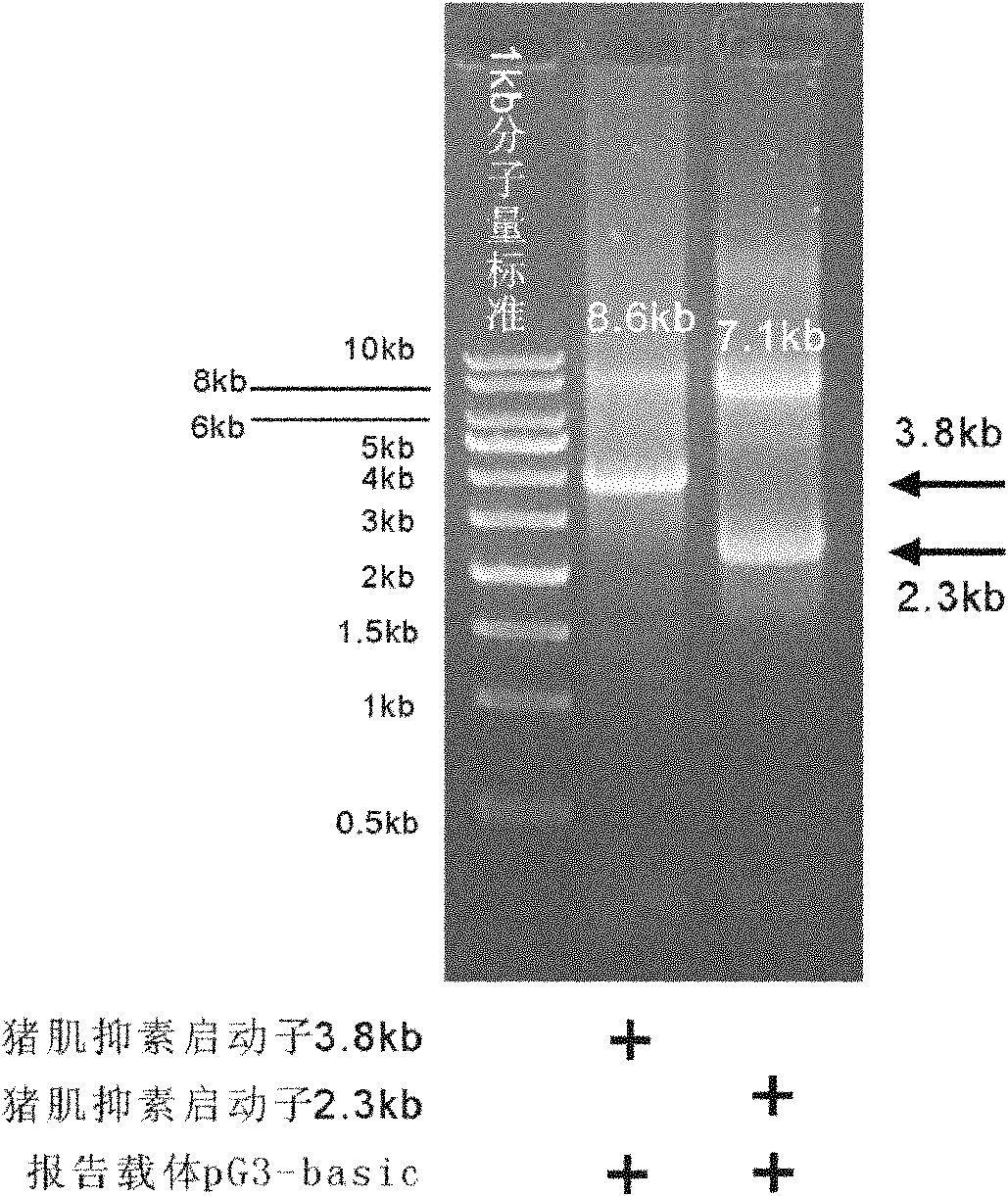
Method for cloning seamless gene
A cloning method and gene technology, applied in the fields of biochemistry and molecular biology, can solve the problems of expensive recombinase, not the preferred solution, and low amplification efficiency, so as to save material cost and time cost, avoid enzyme cleavage connection, simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Design and synthesis of chimeric primers for cloning porcine myostatin promoter.
[0047] The 3.8kb and 2.3b fragments of the 5' end of the porcine myostatin gene were used as its promoter candidate sequence, and were cloned into the 5' end of the firefly luciferase gene of the reporter vector pGL3-basic, so as to facilitate the expression of luciferase by detecting the candidate Candidate sequences are tested for transcriptional activity. According to the working principle of the present invention, chimeric primers are designed respectively for the two candidate sequences (in order to facilitate the distinction, the primers are named according to their length), the sequence is as follows: 3.8kb-pF: 5'gctcgagatctgcgatctaagtaagcttggCATCATTAAACTTCTGACAAGCC3' (SEQ ID No: 1, uppercase Letters represent primers homologous to the 5' end of the porcine myostatin 3.8kb promoter candidate sequence, lowercase letters represent primers homologous to the 5' end of the pGL...
Embodiment 2
[0049] Example 2 One-step binary bridging coupled long-distance PCR amplification of the linear fusion fragment of the porcine myostatin promoter and the reporter vector pGL3-basic.
[0050] i) Sources of reagents and materials
[0051] High-fidelity DNA polymerase KOD plus and its matching buffer (10×buffer), dNTP, magnesium sulfate MgSO 4 All were purchased from Toyobo Biotechnology Co., Ltd., the product number is KOD-201, and stored at -20°C. Pig genomic DNA was extracted according to the conventional phenolic form method and stored at -20°C. The target vector pGL3-basic was carried out according to the instructions of the ultra-pure plasmid extraction kit provided by Beijing Tiangen Biotechnology Co., Ltd., and stored at -20°C.
[0052] ii) System composition: the amplified 3.8kb and 2.3kb fragments are only different in pF primers, and the other components are the same.
[0053] Element
The initial concentration
Dosage
Final concentration
P...
Embodiment 3D
[0061] Embodiment 3Dpn I digests PCR product
[0062] Restriction endonuclease Dpn I specifically recognizes and cuts the methylated GATC sequence, which was purchased from Fermentas Company, Lithuania, Cat. No. ER1702. The composition of reaction system is as follows:
[0063] Element
The initial concentration
Dosage
Final concentration
Buffer Tango TM
10×
3μl
1×
Dpn I
10units / μl
1μl
0.33unit / μl
PCR product
26μl
total capacity
30μl
[0064] Add the above ingredients one by one on ice and mix thoroughly.
[0065] Incubate in a 37°C water bath for 2 hours, and place on ice for transformation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com