Method for identifying foodborne pathogen by surface enhanced Raman spectroscopy
A technology of surface-enhanced Raman for food-borne pathogenic bacteria, applied in biochemical equipment and methods, measurement/inspection of microorganisms, resistance to vector-borne diseases, etc., can solve problems such as time-consuming, high cost, and complicated steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0036] (1) Preparation of microbial samples
[0037] Activation of food-borne pathogenic bacteria strains, after cultivation, the cells were washed in distilled water to make a standard sample of activated food-borne pathogenic bacteria strains, and the samples were injected after completion.
[0038] (2) Optimization of preparation conditions for enhanced reagents
[0039] In the present invention, gold nano-sol, silver nano-sol or other similar reagents can be selected as the reinforcing reagent. Wherein, gold nano-sol and silver nano-sol can be prepared by methods known to those skilled in the art. For example, gold nano-sol can be prepared by reducing chloroauric acid or its salts with sodium citrate, while silver nano-sol can be prepared by reducing soluble silver salt with NaBH4.
[0040] (3) Setting of Raman Spectrometer Injection Conditions
[0041] (4) Collection of Raman spectra of tested bacteria
[0042] The activated bacterial strain standard sample prepared i...
Embodiment 1
[0051]Example 1: Identification of four foodborne pathogens using surface-enhanced Raman spectroscopy: Staphylococcus aureus, Salmonella spp, Listeria monocytogenes and Escherichia coli (Escherichia coli), the process is:
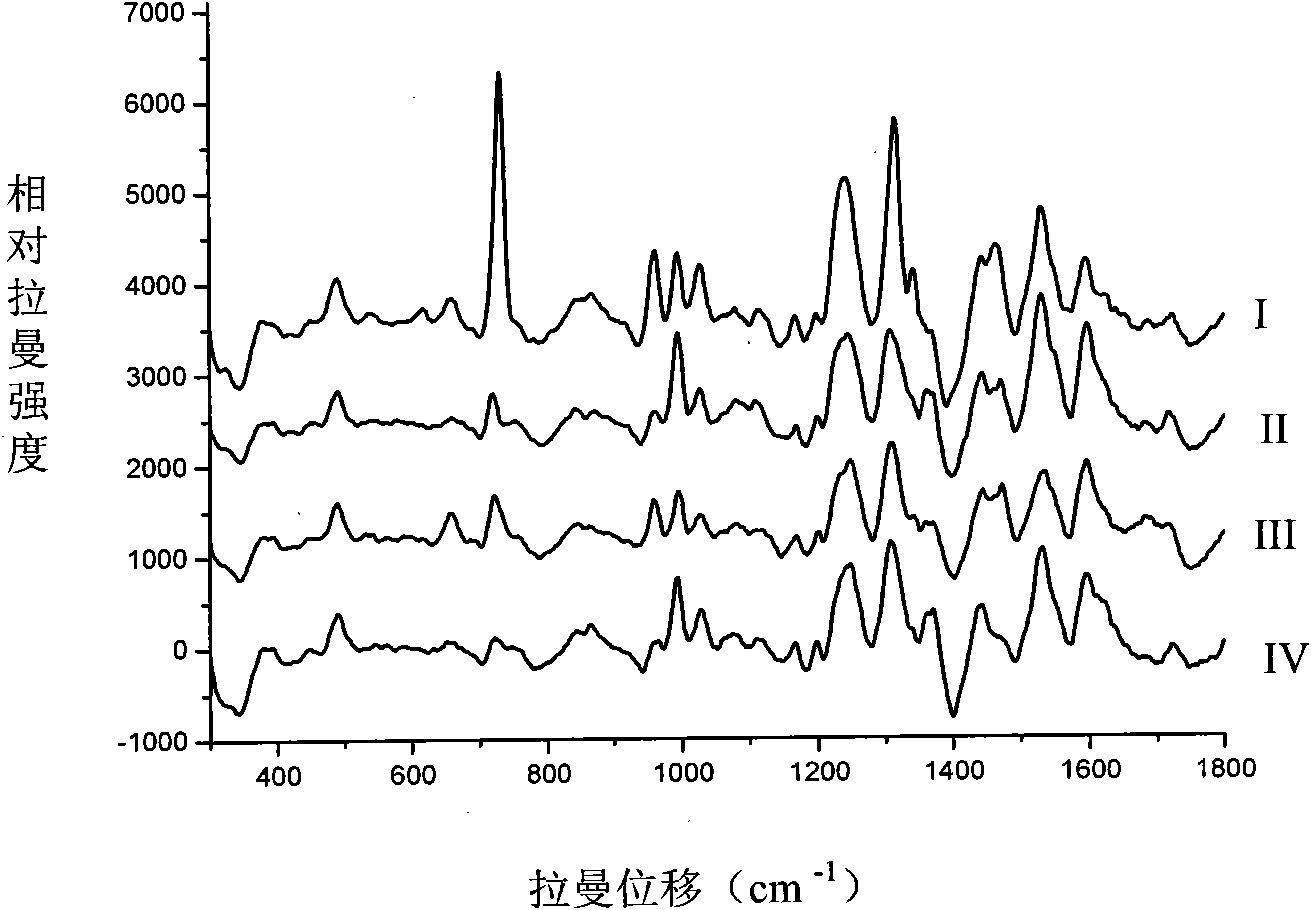
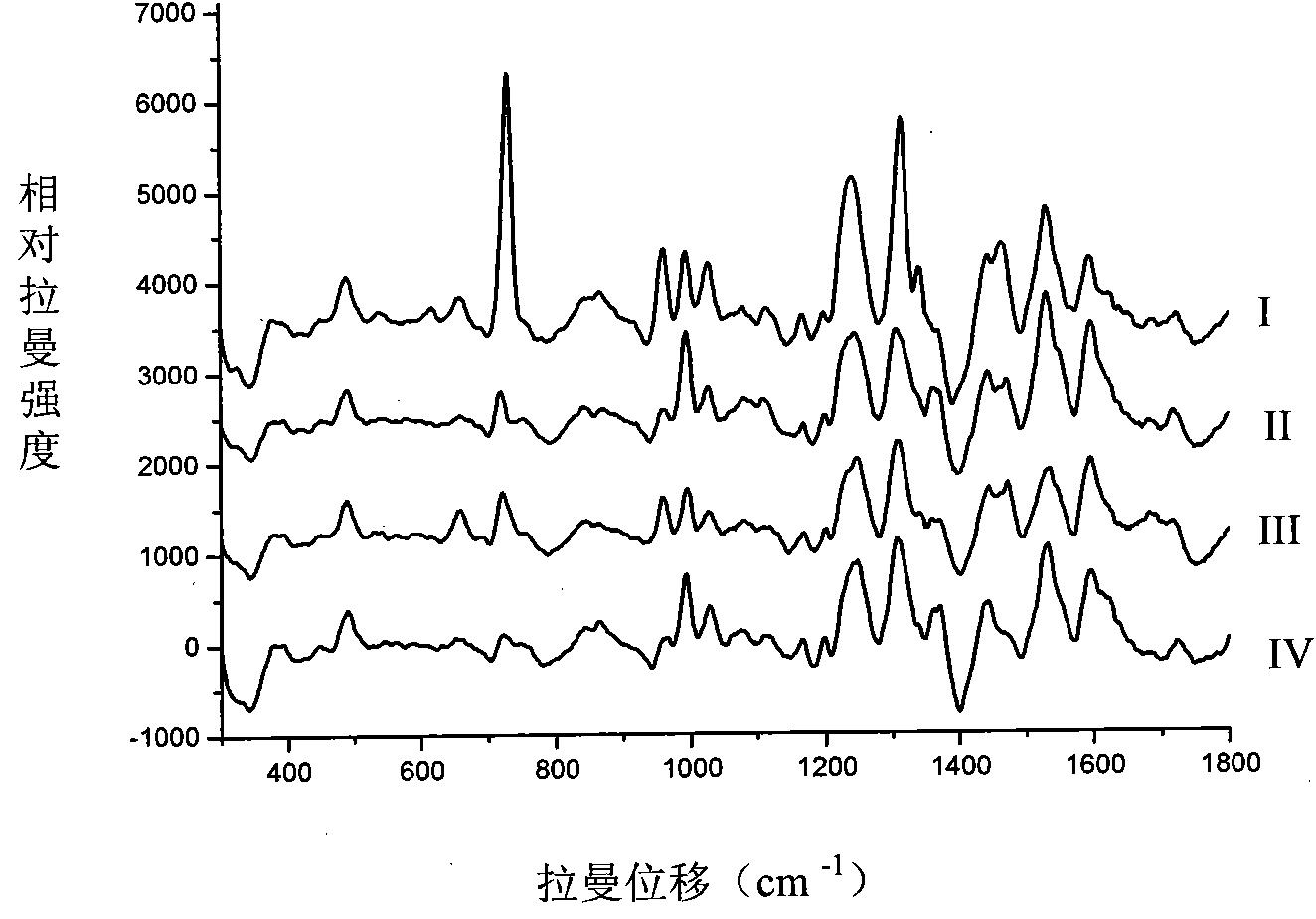
[0052] Take the frozen strains of the above four pathogenic bacteria and inoculate them into TSB centrifuge tubes for activation, and activate them at 30-37°C for at least 24 hours. Scattered colonies were formed after 48 hours. Use a sterile inoculation loop to pick a single colony into a centrifuge tube filled with 4 mL of distilled water, vibrate and turbid, centrifuge in a refrigerated centrifuge at 8500rmp for 5 minutes, and repeat more than 3 times. Take the precipitate and wash it with water before injection. Take 500 μL of the gold nanosol prepared according to the above-mentioned optimization conditions and 300 μL of the above-mentioned bacteria standard sample and mix them in the injection bottle for 8 seconds, and measure 200mw by Raman, and get ...
Embodiment 2
[0053] Example 2: Using Surface Enhanced Raman Spectroscopy to Identify Unknown Foodborne Pathogenic Bacteria Samples
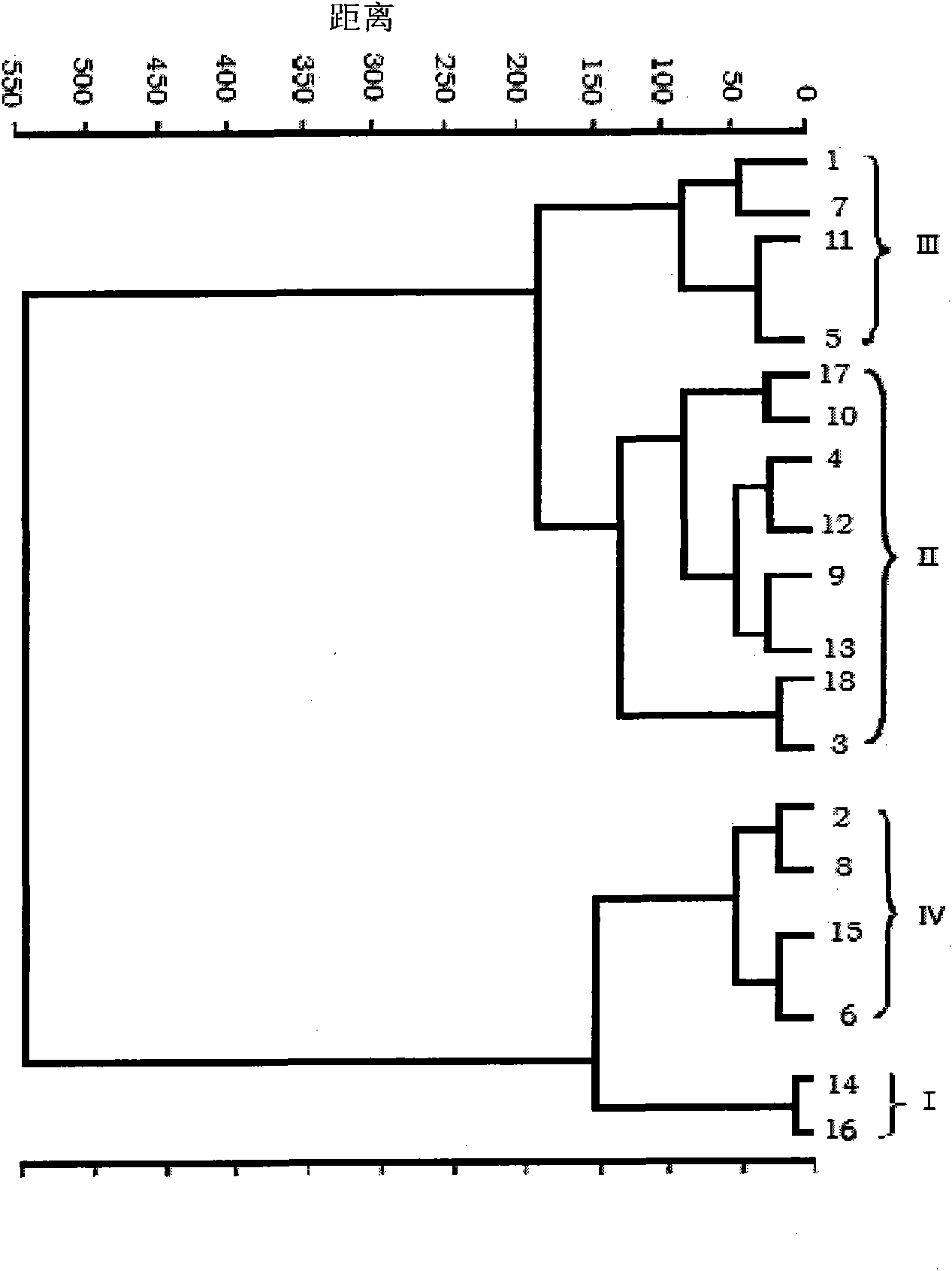
[0054] In this embodiment, according to the above method, 18 unknown food-borne pathogenic bacteria samples were sampled separately, and the obtained Raman spectrum was collected, and the spectral data were processed by SPSS software to obtain the cluster analysis results, and then the spectral data and cluster analysis The results were compared with the spectrograms and cluster analysis results of the aforementioned standard samples, and thus determined samples numbered 14, 16, samples 3, 4, 9, 10, 12, 13, 17, 18, 1, The samples of 5, 7, and 11, and the samples of 2, 6, 8, and 15 were Listeria monocytogenes, Salmonella, Escherichia coli, and Staphylococcus aureus, respectively, thus realizing the unknown food-borne pathogenic bacteria sample species Genus identification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com