Construction and application of URA3 defective P. pastoris X-33 strain
A defect-type, strain-based technology, applied in the field of bioengineering, can solve problems such as the inability of yeast to survive, and achieve the effect of stabilizing genetic traits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
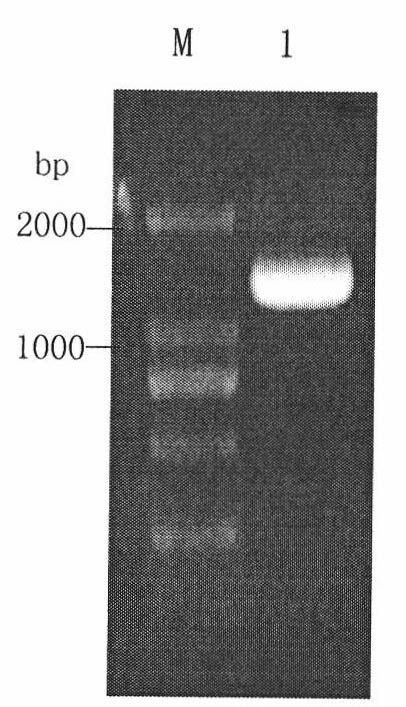
[0018] Cloning of URA3 gene and construction of homologous replacement DNA fragments: URA5', URA3' and fusion homologous arm fragments at both ends of URA3 gene PCR amplified from Pichia pastoris X-33 genome were 0.7kb, 0.7kb, 0.6kb and 1.3kb ( figure 1 ). Sequencing results showed that each sequence and fusion sequence were correct.
Embodiment 2
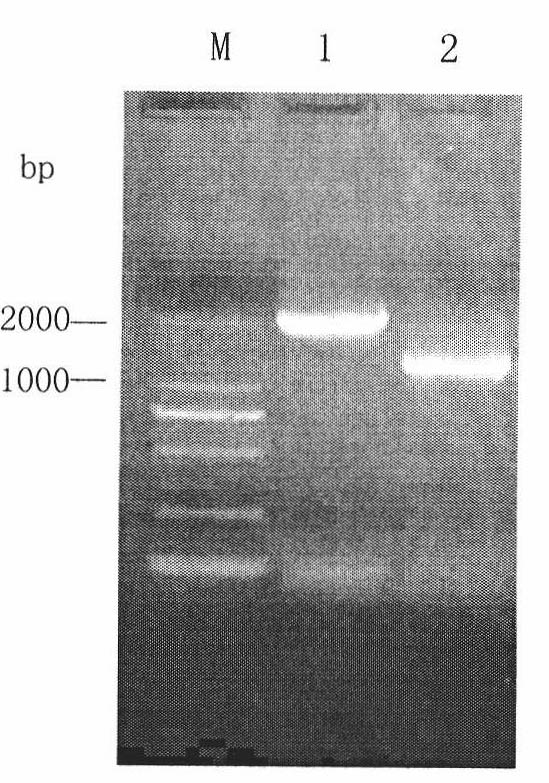
[0020] Knockout and identification of URA3 gene: After the URA5'-3' fusion homology arm fragment was electroporated into yeast, double-crossover homologous recombination occurred with the URA3 gene in the genome, replacing about 700 bp of the sequence in the coding frame of the URA3 gene. The strains without homologous recombination cannot grow on the medium containing 5-FOA because they carry the URA3 gene, so the ura3 deletion strain can be screened on the MD medium containing 5-FOA and uracil. Select strains that have been phenotype-identified and have stable traits, and use external primers URA5F and URA3R for genomic PCR identification. The size of the URA3 gene of wild-type X-33 bacteria is 2043bp, so the amplified product is about 2000bp; the URA3 gene encoding of ura3-deleted bacteria The sequence of about 700bp in the box was replaced, and the amplified product should be about 1300bp, and the electrophoresis results were consistent with expectations ( figure 2 ), ind...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com