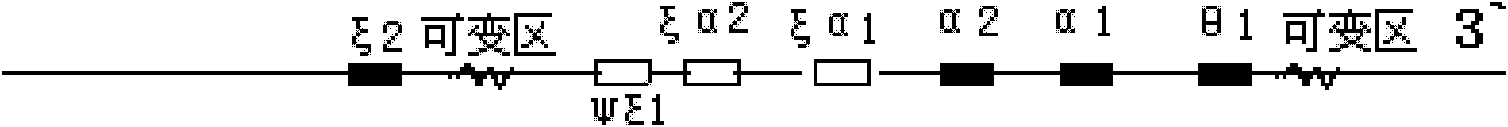Method and kit for detecting southeast Asia deletion alpha-thalassemia
A technology of thalassemia and detection method, which is applied in the field of detection of deletional α-thalassemia in Southeast Asia, which can solve the problems of long experimental period, unfavorable clinical promotion and application, and cumbersome operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Synthesis and purification of primers and probes
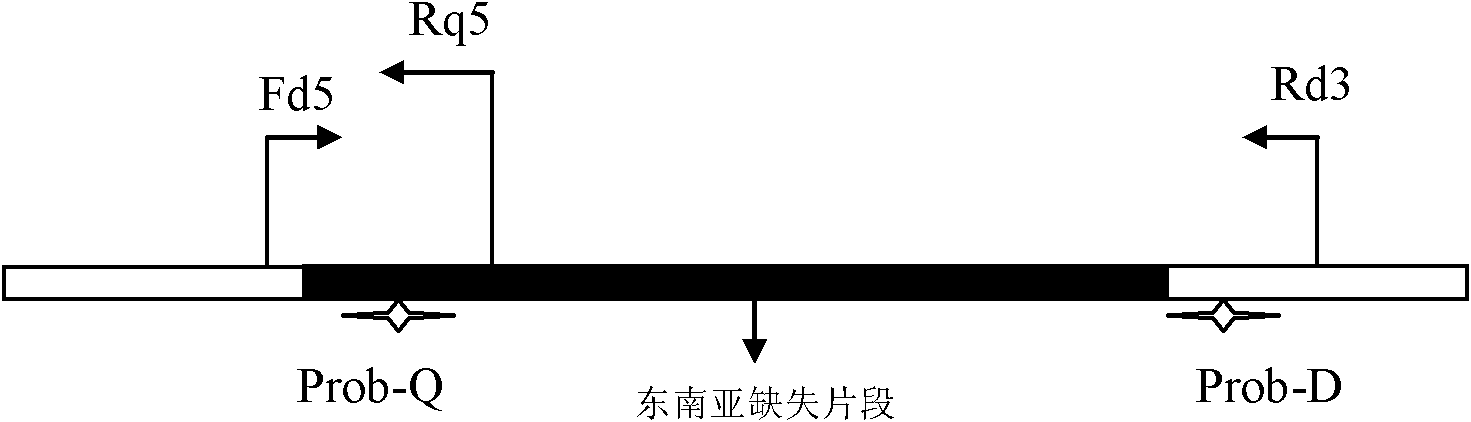
[0056] Using the α-globin gene with Genebank accession number AE006462 as the target sequence for PCR amplification, the following primers were designed and synthesized:
[0057] Fd5: the nucleotide sequence shown in SEQ ID NO: 1;
[0058] Rd3: the nucleotide sequence shown in SEQ ID NO: 2;
[0059] Rq5: the nucleotide sequence shown in SEQ ID NO: 3;
[0060] The following Taqman probes were synthesized according to the sequence design between the primers:
[0061] Prob-D: 5'-VIC-TTGGCCAGGTGCCGTGGCTT-HBQ-3';
[0062] Prob-Q: 5'-FAM-CCGGGATTCGGACGTCAGGC-HBQ-3'.
[0063] The synthesized primers were purified by PAGE, and the synthesized probe sequences were purified by HPLC. The fluorescent probe Prob-Q uses FAM as the reporter fluorescence; HBQ is used as the quencher; the fluorescent probe Prob-D uses VIC as the reporter fluorescence; HBQ is used as the quencher.
Embodiment 2
[0064] Example 2: Primer Specificity and Sensitivity Analysis
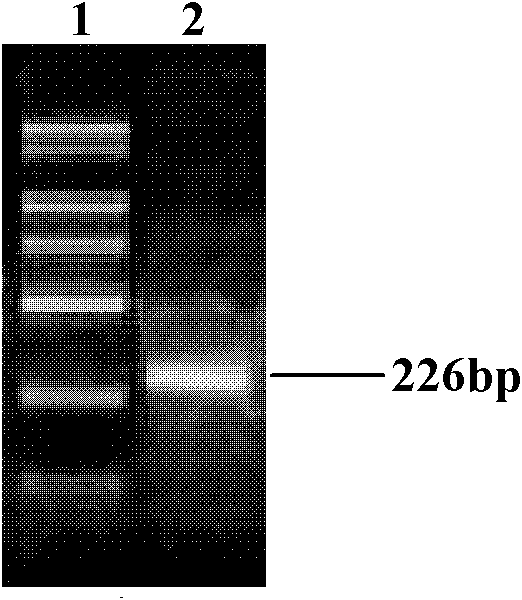
[0065] Using the Southeast Asia deletion homozygous genome as a template to carry out PCR amplification with the primer set having the primer Fd5 of the nucleotide sequence shown in SEQ ID NO: 1 and the primer Rd3 having the nucleotide sequence shown in SEQ ID NO: 2 , for gel electrophoresis detection see image 3 , using the normal human genome as a template to carry out PCR amplification with the primer set of the primer Fd5 having the nucleotide sequence shown in SEQ ID NO: 1 and the primer Rq5 having the nucleotide sequence shown in SEQ ID NO: 3, gel Electrophoresis detection see Figure 4 . Depend on image 3 and Figure 4 As shown, the gel electrophoresis detection of primers Fd5 and Rd3 and the amplification products of primers Fd5 and Rq5 were all single bands, and fewer primer dimers were generated during amplification, indicating good primer specificity.
[0066] The amplified products of primers Fd...
Embodiment 3
[0068] Embodiment 3: Construction of reference plasmid
[0069] 1. Preparation of normal human gene amplification product plasmid control substance:
[0070] Collect normal samples, and use commercial whole blood genome purification reagents to extract the human genome in normal samples. A normal human genome sample is amplified with a primer pair of Fd5 and Rq5, and the obtained PCR product is cloned into a T-vector plasmid. The plasmid was transformed into Escherichia coli JM109, screened and cultivated in LB medium containing ampicillin, and a single clone was selected and cultured by shaking. Extract the plasmid, after purification, detect the concentration, calculate the copy number, and use it as a normal human gene amplification product plasmid reference substance for future use.
[0071] 2. Preparation of plasmid reference substance for homozygous gene amplification product of Southeast Asia deletion:
[0072] Samples of deletional α-thalassemia in Southeast Asia we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com