M-PCR (Multiplex Polymerase Chain Reaction) primers, probes and detection methods for vibrio cholerae, vibrio parahaemolyticus and salmonella
A technology for hemolytic Vibrio and Vibrio cholerae, which is applied in the field of detection of pathogenic microorganisms in food, can solve the problems of multiple PCR technology and achieve high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, establishment of detection method
[0035] 1. Design of primers and probes
[0036] According to the results of homologous gene screening comparison, the fimY gene (SEQ ID NO.10) of Salmonella, the hlyA gene (SEQ ID NO.11) of Vibrio cholerae and the toxR of Vibrio parahaemolyticus were selected with high specificity The gene (SEQ ID NO.12) is the target gene for detecting amplification. According to the nucleic acid sequences of the above-mentioned genes published on NCBI, and through comprehensive consideration of factors such as the consistency of the annealing temperature of the detection primers and probes and the similarity of GC content, the following specific primers and probes were designed:
[0037] ① Detection primer and probe sequence of fimY gene of Salmonella:
[0038] Forward primer (SEQ ID NO.1): 5'-CCGTATGGCTGGGCGTTT-3',
[0039] Reverse primer (SEQ ID NO.2): 5'-AGTACGGCTAAAGCTTTCCGATAAG-3',
[0040] Probe (SEQ ID NO.3): 5'FAM-CAGAGGCCA...
Embodiment 2
[0070] Embodiment 2, specificity test
Embodiment 3
[0078] Embodiment 3, sensitivity test
[0079] 1. Detection sensitivity of pure culture of standard strains
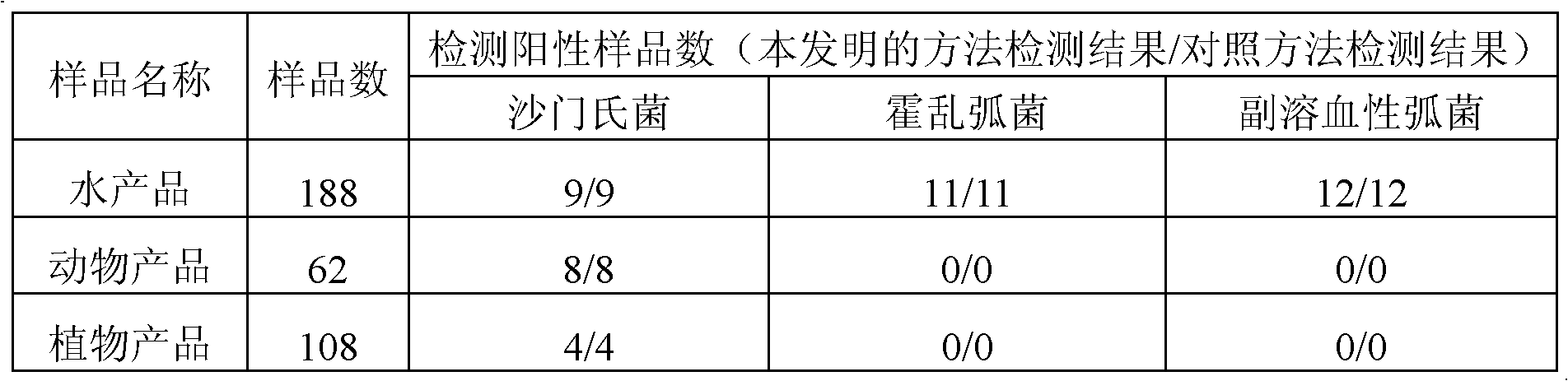
[0080] Using the specific primers, probes and detection methods of Example 1, the detection sensitivity of pure cultures of Salmonella, Vibrio cholerae and Vibrio parahaemolyticus standard strains was studied.
[0081] Take the above three standard strains and culture them in ordinary broth medium for about 18 hours, measure their OD value, estimate the number of bacteria, and then carry out gradient dilution to 10 times in ten-fold increments. -7 , take the bacterial solution of the last four dilutions for plate counting, repeat 3 times for each dilution, perform colony counting and take the average value after culturing according to the corresponding method to determine the true bacterial density. Simultaneously each dilution respectively gets 1mL bacterium liquid in 2mL centrifuge tube and makes 3 tubes to repeat, is used for extracting DNA and applies the method o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com