Kit for in-vitro single base damage repair detection through real-time quantitative PCR (Polymerase Chain Reaction)
A damage repair and in vitro detection technology, applied in the field of DNA single base damage repair detection, can solve the problems of restricted environment, human injury, pollution, etc., and achieve the effect of ensuring specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] First: Template Annealing:
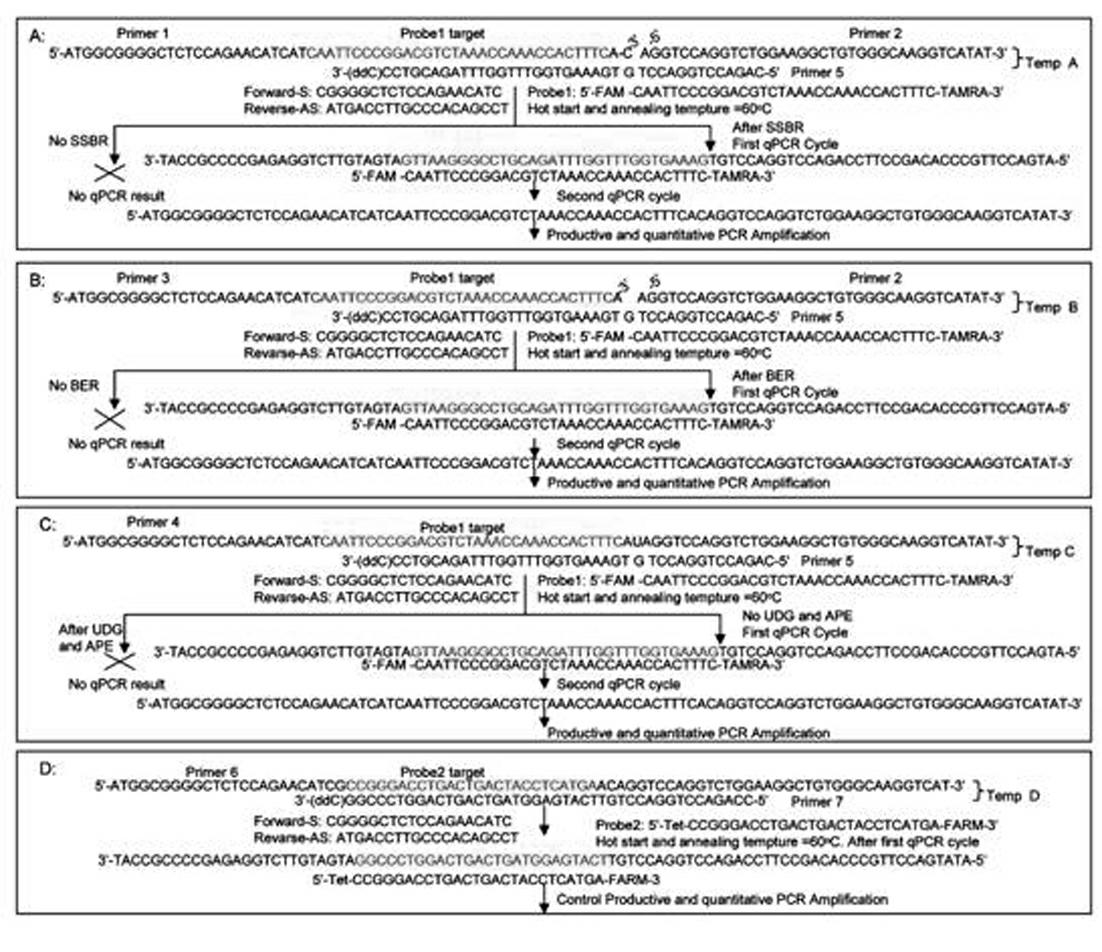
[0047]Take 200 pmol of oligonucleotides (Primer1-7) shown in SEQ ID No.1 to SEQ ID No.7 respectively, combine them according to Table 1 and add them to four EP tubes respectively. Table 1 is templates A, B, C, D. Template A consists of Primer1 (SEQ ID No.1), Primer2 (SEQ ID No.2) and Primer5 (SEQ ID No.5); Template B consists of Primer3 (SEQ ID No.3), Primer2 (SEQ ID No.2) ) and Primer5 (SEQ ID No.5); Template C is composed of Primer4 (SEQ ID No.4) and Primer5 (SEQ ID No.5); Template D is composed of Primer6 (SEQ ID No.6) and Primer7 (SEQ ID No.5) No.7) composition. Add 20ul of 10x annealing buffer (250mM NaCl; 20 mM Tris-Cl, pH=8.0; 10mM EDTA) to four EP tubes, make up to 200ul with water, anneal at 72°C for 10 minutes, then gradually drop to room temperature. Store at -20°C for later use.
[0048] Table 1 Template A, B, C, D combination
[0049]
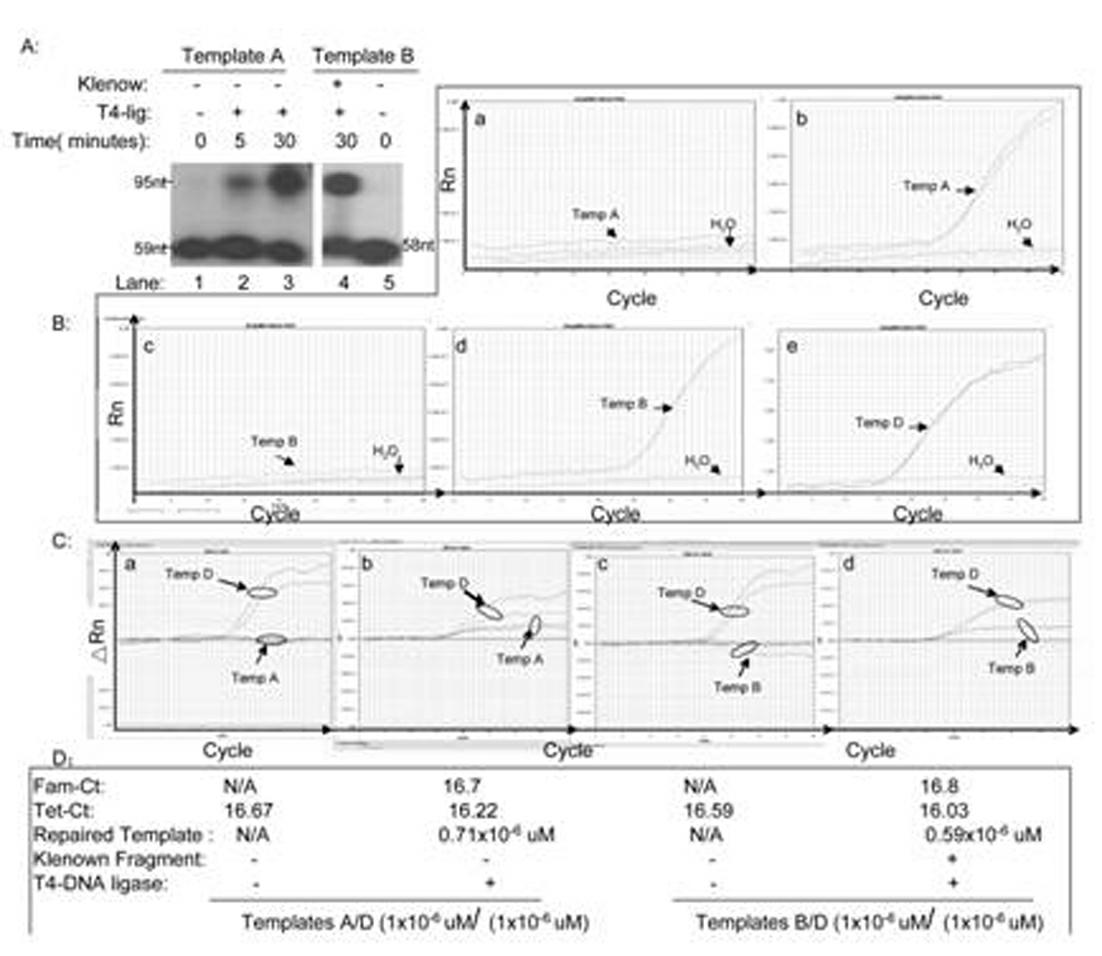
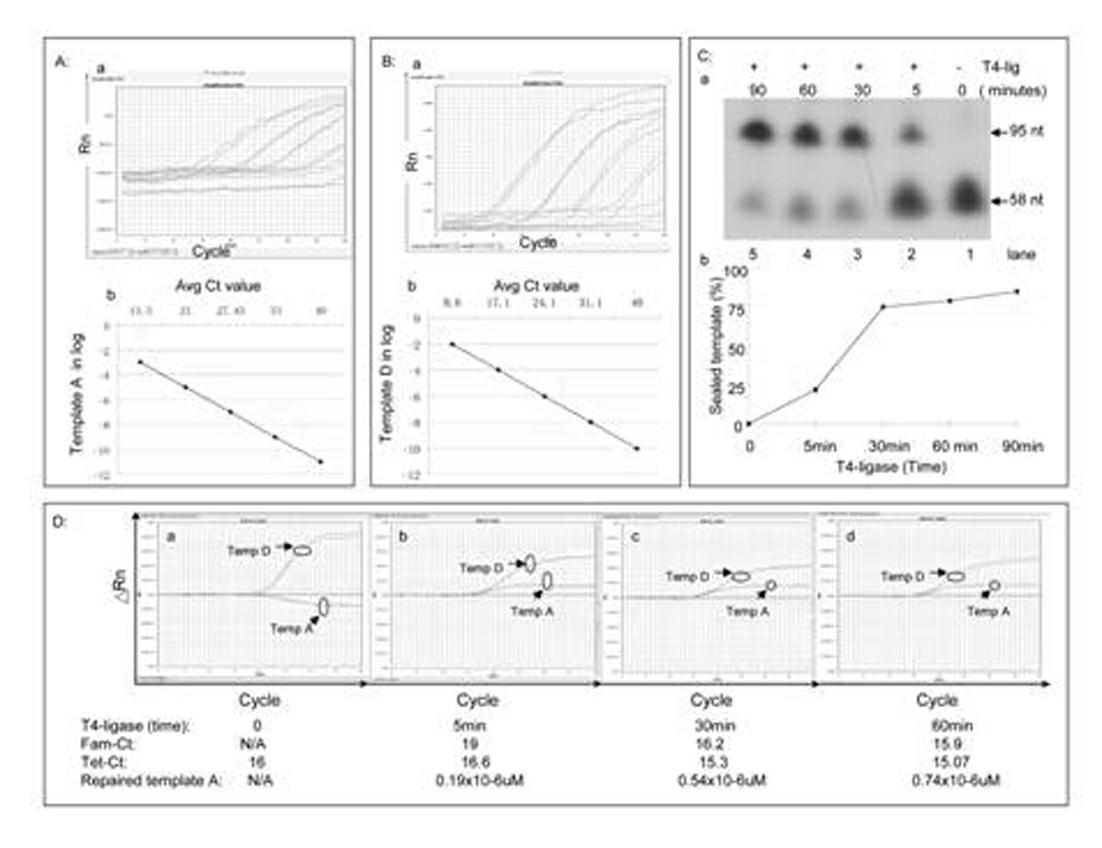
[0050] Second: single base deletion damage repair ability detection and sensitivity a...
Embodiment 2
[0065] Example 2 Validation of the kit and detection method of the present invention
[0066] In the usual in vitro single-base damage repair experiments, people use isotopes 32 P-labeled damaged oligonucleotide substrates and purified enzyme and nucleotide extracts to determine DNA polymerase, DNA ligase and glycosidase activities. However, due to the application of isotopes, the study of single-base damage repair in vitro is greatly limited. In this research mode, the templates include: single-strand break template A, single-nucleotide removal template B, single-base modified oligonucleotide template C and control oligonucleotide template D, specifically as follows figure 1 shown. In this novel qPCR method, we designed two different target sequence probes for modified and control oligonucleotides. The modified oligonucleotide target probes were labeled with 5'-FAM, and the control oligonucleotide target probes were labeled with 5'-Tet. All oligonucleotides (Primer 1 to 7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com