Detection method for BRAF gene mutation
An allele and sequence technology, applied in the field of molecular biology method to detect mutant bases, can solve the problems of difficult to meet clinical detection, long detection period, easy pollution, etc., to improve the accuracy and detection efficiency, and the steps are simple. , the effect of saving time and cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
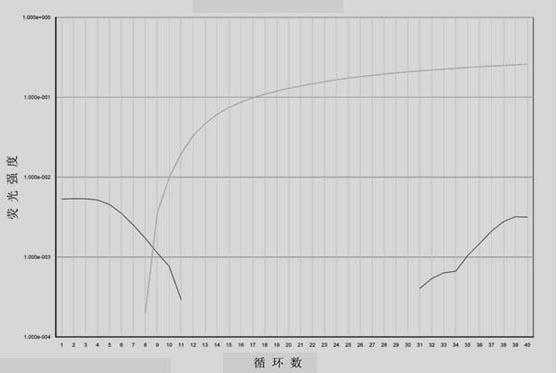
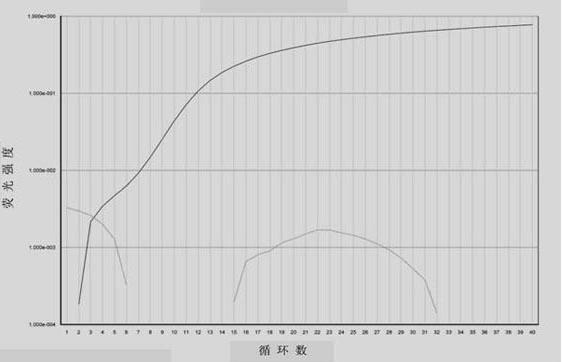
[0030] 1. Design and synthesis of BRAF gene-specific amplification primers and taqman-MGB probe:
[0031] According to the position and genotype of the mutation site, a pair of gene-specific upstream and downstream amplification primers (the length of the amplification product is 136bp) and taqman-MGB probes with different fluorescein labels for wild-type and mutant BRAF genes were designed. One needle each. Primers and probes were synthesized by Shanghai Jikang Biotechnology Co., Ltd. The specific sequence is as follows.
[0032] BRAF amplification primers:
[0033] BRAF-F1 (forward): 5' CTTACCTAAACTCTTCATAATGCTTGC 3'
[0034] BRAF-R1 (reverse): 5' TAGCCTCAATTCTTACCATCCACA 3'
[0035] BRAF gene wild-type probe: 5'-E-AGCTACAGTGAAATC-P-3'
[0036] BRAF mutant probe: 5'-F-AGCTACAGAGAAATC-P-3'
[0037] Where: E=HEX reporter dye, F=FAM reporter dye, P=Taqman-MGB group
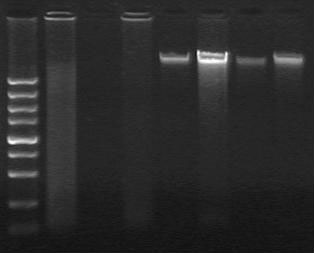
[0038] 2. Extraction of tumor tissue DNA:
[0039] (1) Sample collection: Take 2 cases of fresh tissue b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com