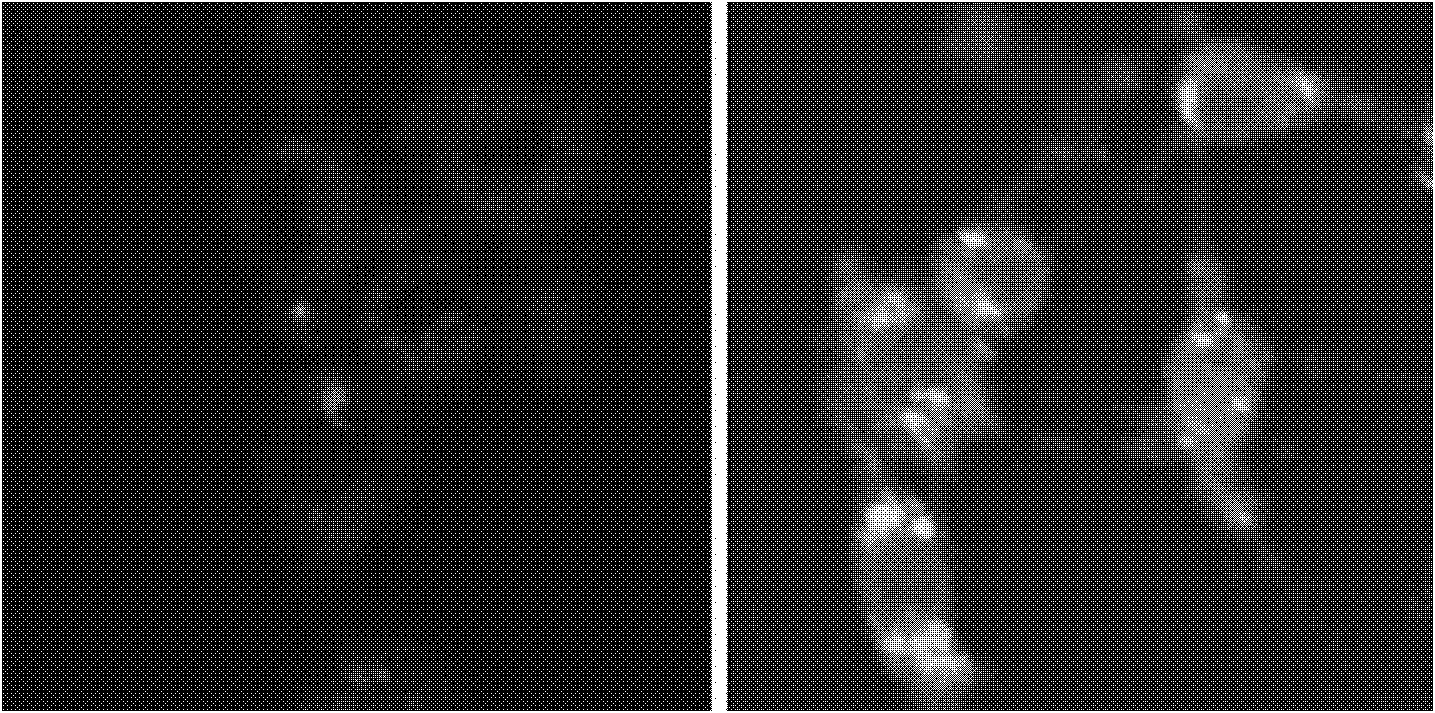Probe for detecting nucleic acid in living cells and application method thereof
A detection method and technology for living cells, applied in the nanotechnology and biological fields, can solve the problems of not suitable for in-situ detection of living cell samples, cumbersome operations, not fully applicable to living cell samples, etc., and achieve real-time in-situ detection and enhanced detection. Sensitivity, the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
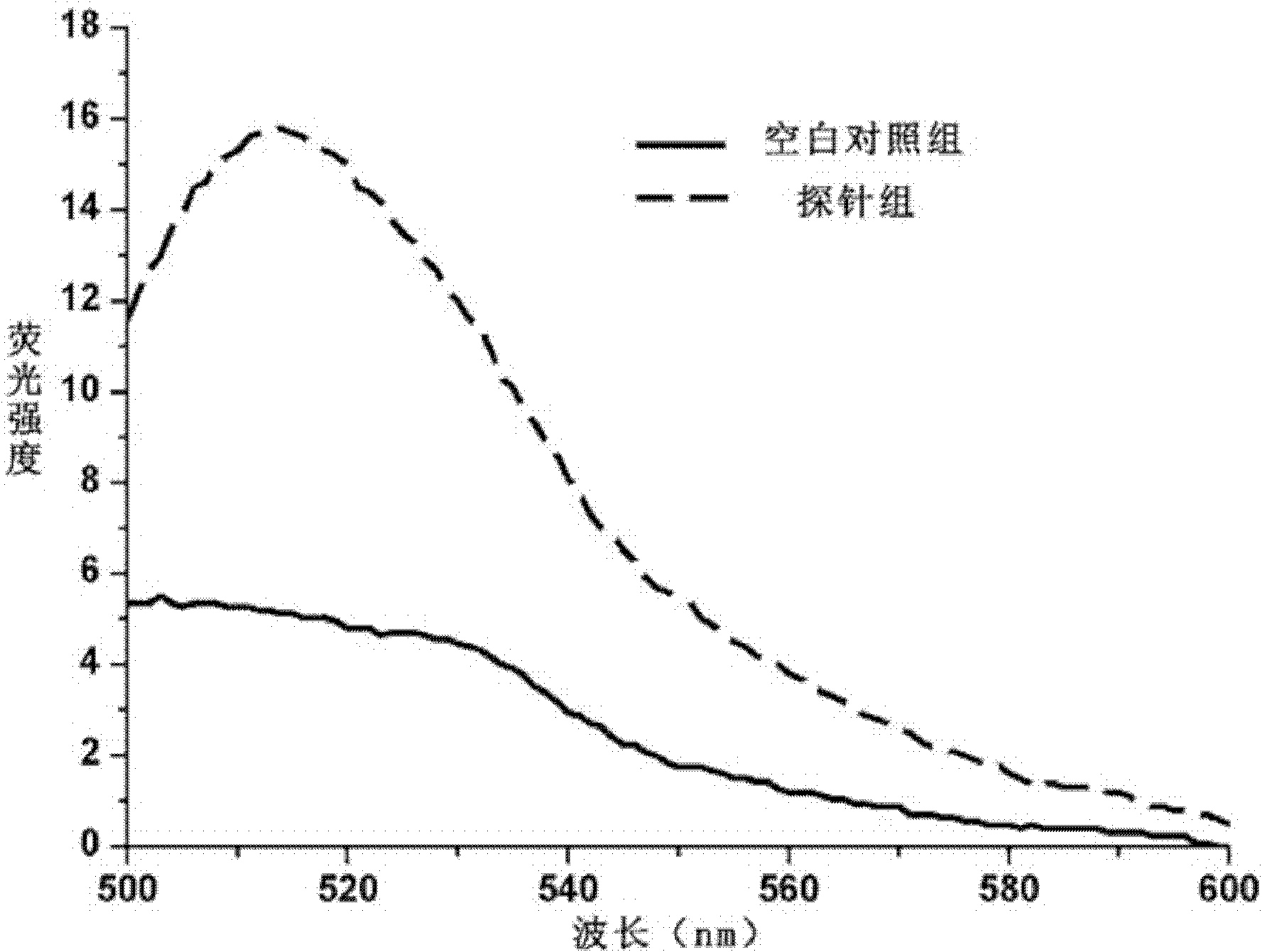
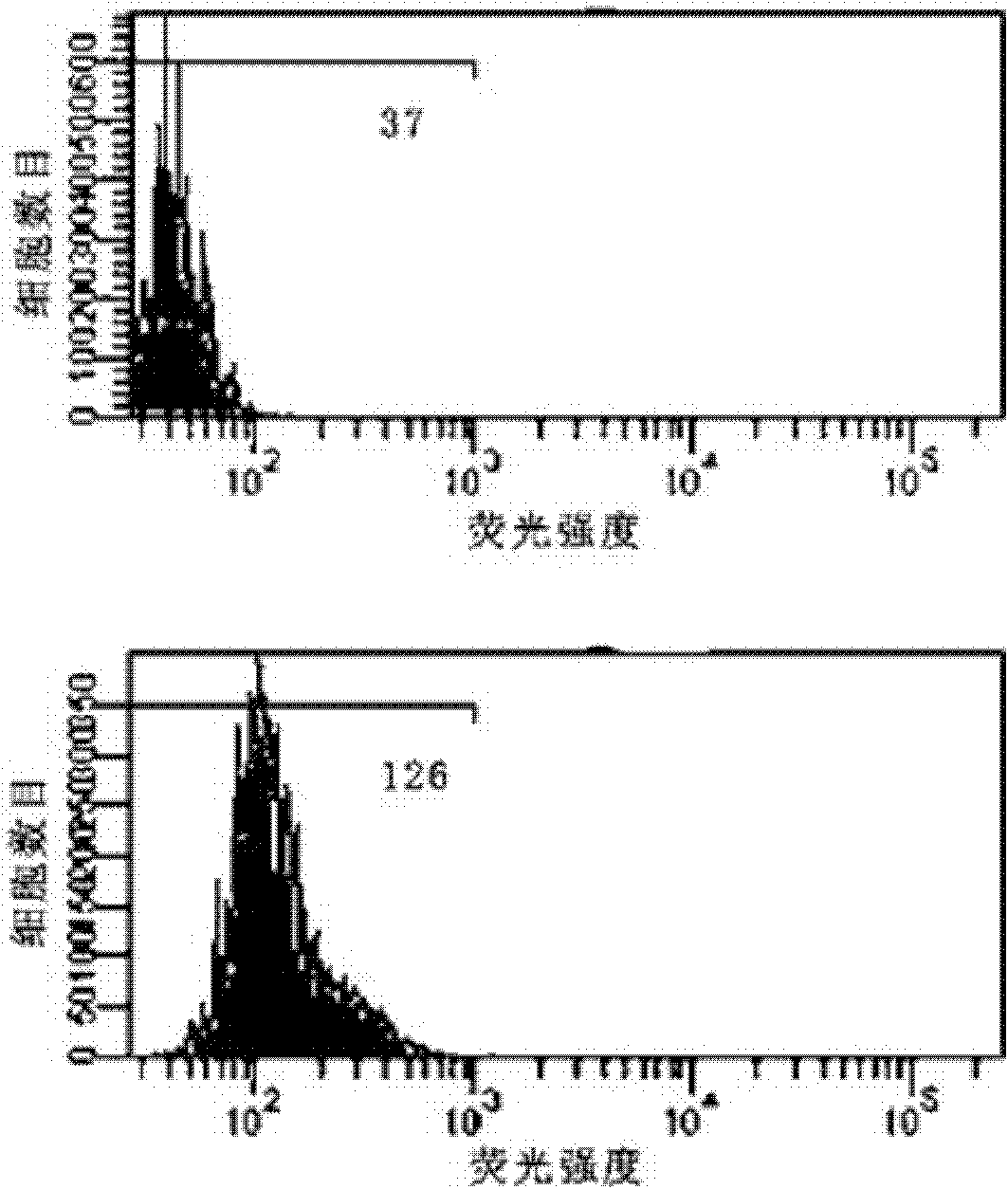
[0045] Signal transducer and activator of transcription 5B (STAT5B) is related to various diseases such as breast cancer, prostate cancer and leukemia, so detecting the expression of STAT5B mRNA (SEQ ID NO: 1) is helpful for early diagnosis of tumors.
[0046] Preparation of the probe: For the mRNA synthesis probe of human STAT5B, the hairpin oligonucleotide (SEQ ID NO: 2) and the target sequence (SEQ ID NO :3), the 3' end of the hairpin targeting nucleic acid was labeled with FITC, and the 5' end was modified with a sulfhydryl group. In phosphate buffered saline (PBS, 0.02mol / L, pH=8.0), fluorescent substance-labeled and sulfhydryl-modified hairpin oligonucleotides were mixed with dithiothreitol (DTT) at a molar ratio of 1:10 for 45 Minutes, and then repeated 4 times with 4 times the volume of ethyl acetate to extract the DTT in the reaction. Then add the above reaction solution dropwise into the gold nano solution at a molar ratio of 100:1, mix well, react for 24 hours, add...
Embodiment 2
[0052] Clinical studies have shown that ERCC1-negative patients have better curative effect after using cisplatin, while ERCC1-positive patients have no obvious effect, so ERCC1 gene is a drug-sensitive gene. Therefore, detecting the expression of ERCC1 can guide the clinical medication of tumor therapy.
[0053] Preparation of the probe: Synthesize the probe against ERCC1 mRNA (SEQ ID NO: 4), please use the hairpin oligonucleotide (SEQ ID NO: 5) and target Sequence (SEQ ID NO: 6), the 3' end of the hairpin targeting nucleic acid was labeled with FITC and the 5' end was modified with a sulfhydryl group. In phosphate buffered saline (PBS, 0.02mol / L, pH=8.0), fluorescent substance-labeled and sulfhydryl-modified hairpin oligonucleotides were mixed with dithiothreitol (DTT) at a molar ratio of 1:10 for 45 Minutes, and then repeated 4 times with 4 times the volume of ethyl acetate to extract the DTT in the reaction. Then add the above reaction solution dropwise into the gold nan...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com