Method for producing high-activity and high-purity rifamycin SV
A technology of rifamycin and cytochrome, applied in the field of using the bacteria, can solve the problems such as not very clear
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118] Example 1: Whole-genome sequencing of the rifamycin Rf_SV strain U32 and its sequence comparison with the rifamycin B strain
[0119] The rif16 gene was sequenced by the inventors after sequencing the entire genome of a strain of A. mediterranei strain U32 producing rifamycin Rf_SV and comparing it with three wild-type strains producing rifamycin B. discovered during the process. in particular:
[0120] Using the second-generation 454 sequencer, combined with SOLID, Sanger sequencing methods, and 6-8k plasmid library and fosmid library construction, the complete gene sequence of 10,236,715bp Amycobacterium mediterranei U32 with an error value of less than 0.5 / 100,000 was finally obtained . After the rifamycin synthesis gene cluster rif was obtained by BLASTP annotation, it was compared with the rif synthesis gene cluster of the Rf_B-producing strain S699 studied by Floss. The inventors discovered 41 SNPs and 8 INDEL sites, which altogether affected the promoter regi...
Embodiment 2
[0130] Embodiment 2: transfer rif16 of ATCC 21789 to U32
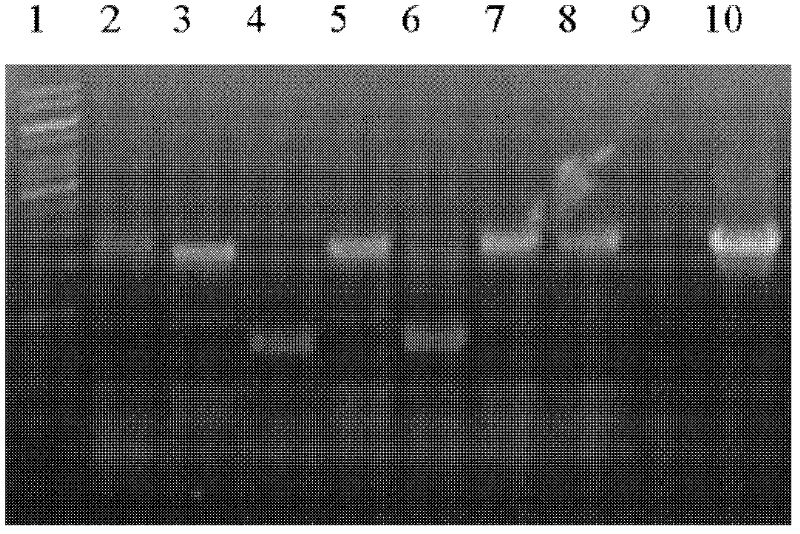
[0131] Using conventional molecular biology methods, the PCR product of the rif16 gene containing its own promoter region in ATCC 21789 (the primers used are shown in SEQ ID Nos: 19 and 20) was cloned into the episomal multi-copy plasmid of A. mediterranei On the EcoR V site of pDXM4 (refer to Ding Xiaoming 2001), a plasmid pDXM4-P450 containing wild-type ATCC 21789 rif16 gene was obtained. Using the empty plasmid as a control, pDXM4-P450 containing rif16 was transferred to U32 by electroporation (refer to Ding Xiaoming 2001). The result is as Figure 5 shown.
Embodiment 3
[0132] Embodiment 3: The stability of U32 complementation plasmid pDXM4-P450 is transformed into the influence of Rf_B by Rf_SV
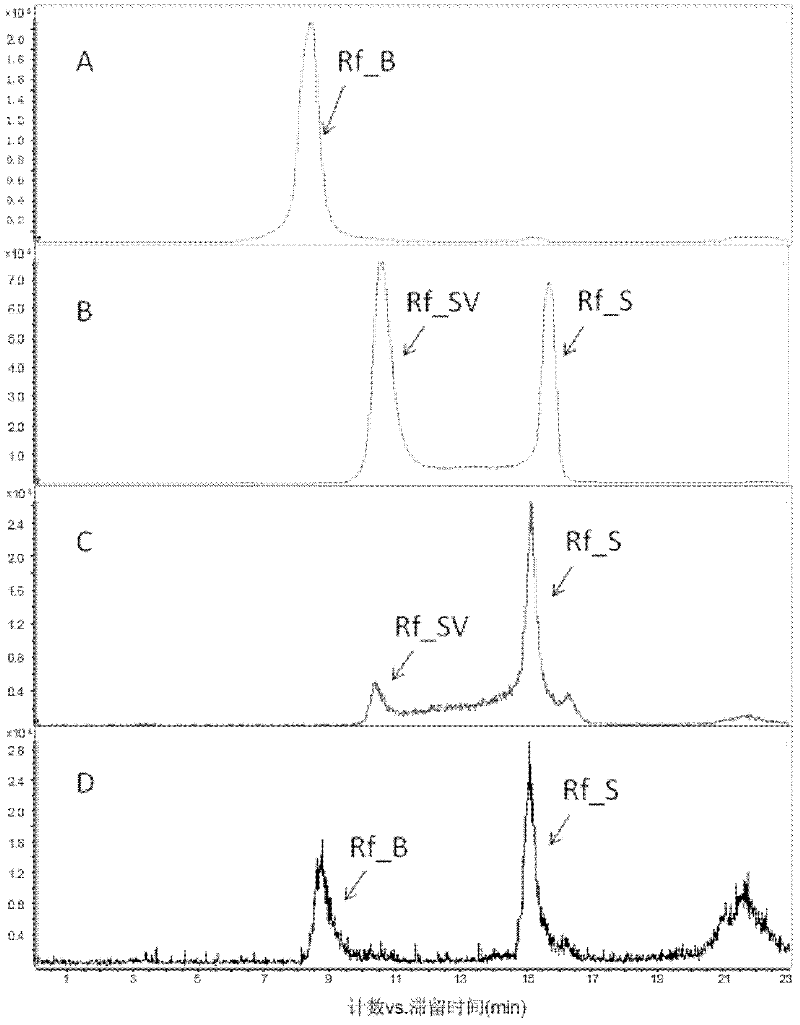
[0133] The plasmid pDXM4-P450 can be stabilized in the U32 strain if the antibiotic apramycin is added to the medium, otherwise the plasmid will be lost during the proliferation of the bacteria. Taking the addition of apramycin antibiotics in the medium as a control, we took samples at the time points of 2 days, 3 days, and 5 days in the U32 fermentation broth without adding apramycin antibiotics, and did it together with the fermentation broth with antibiotics added. HPLC-MS detection. The results found that with the loss of the complementing plasmid pDXM4-P450, Rf_SV and Rf_S in the fermentation broth accumulated rapidly. It is verified from the side that rif16 plays a role in the conversion of Rf_SV to Rf_B ( Figure 6 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com