Lycopene Cyclase Gene of Sphingomonas paucimobilis and Its Application
A technology of lycopene and less active sheath, which is applied in the fields of application, genetic engineering, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1: Obtaining of DNA sequence of crtY gene of Sphingomonas paucimobilis
[0021] According to the partial sequence of Sphingomonas paucimobilis ATCC 31461 crtY (NCBI accession number: HQ202920), specific primers for upstream genes were designed, and the complete sequence including crtY gene and the sequence with NCBI accession number HQ202920 were obtained by SiteFinding-PCR technology Part of the flanking sequence required for gene knockout was obtained by splicing. The sequences of the upstream gene-specific primers, SiteFinder, and SFP primers used are as follows:
[0022] Upstream sequence-specific primers:
[0023] UP1: 5′-GCTGTAATAGGTGTCCTCGACGA-3′
[0024] UP2: 5′-TGAACGGCAGGCAATAGACGAAG-3′
[0025] SiteFinder:
[0026] SiteFl:
[0027] 5′-CACGACACGCTACTCAACACACCACCACGCACAGCGTCCTCAANNNNNNCATGG-3′
[0028] SiteF2:
[0029] 5′-CACGACACGCTACTCAACACACCACCACGCACAGCGTCCTCAANNNNNNCATGC-3′
[0030] SiteF3:
[0031] 5′-CACGACACGCTACTCAACACACCACCACGCACAG...
Embodiment 2
[0038] Example 2: Construction of a recombinant strain of Sphingomonas paucimobilis deficient in lycopene cyclase
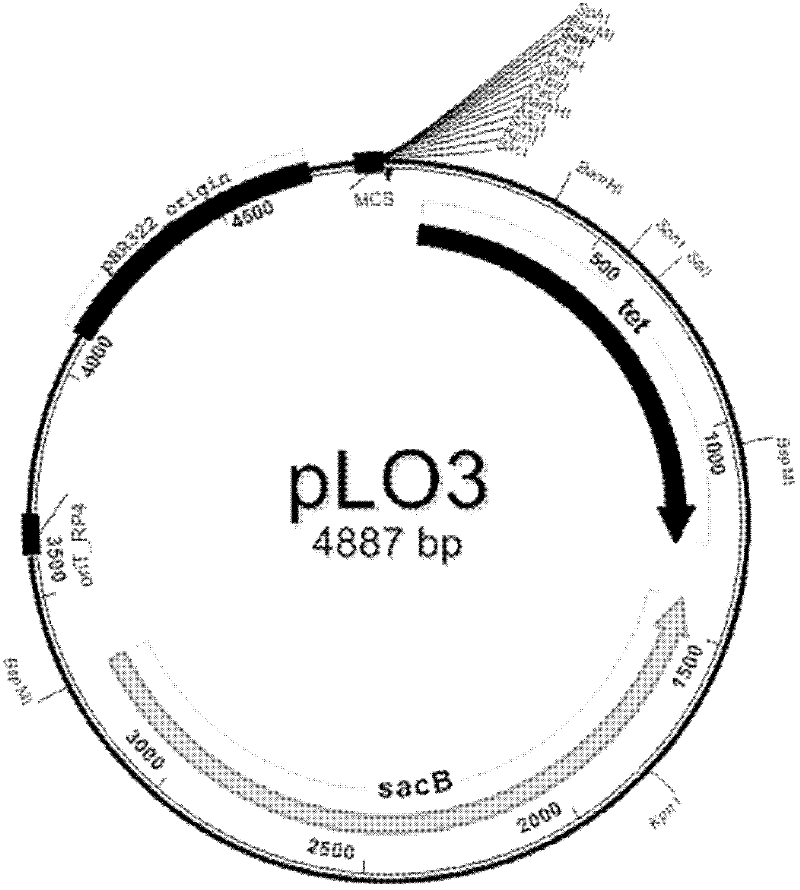
[0039] 1. Construction of gene knockout vector pLO3-△Y
[0040] Primers were designed based on the obtained DNA sequences. Genomic DNA (using the AxyPrep Bacterial Genomic DNA Miniprep Kit) was extracted from Sphingonmonas paucimobilis ATCC 31461 (purchased from ATCC) as a PCR template. Y1 and Y2 are primers Taq enzyme (Takara) to amplify upstream homologous sequences. The PCR reaction program is 95°C pre-denaturation for 5 minutes to enter the cycle process; Extend at 72°C for 10 min. Y3 and Y4 are the primers Taq enzyme (Takara) to amplify downstream homologous sequences. The PCR reaction program is: 95°C pre-denaturation for 5 minutes to enter the cycle process; 94°C denaturation for 30 sec, 63°C annealing for 30 sec, 72°C extension for 40 s, 30 cycles, Finally, it was extended at 72°C for 10 min. The above primer sequences are as follows:
[0041] Y1: 5′-A...
Embodiment 3
[0064] Example 3: Determination of Gel Production Ability of Sphingomonas paucimobilis Strains Deleted in crtY Gene
[0065] 1. Wild-type strain (Sphingomonas paucimobilis ATCC 31461) and crtY gene-deleted Sphingomonas paucimobilis (△Y1~△Y7, where △Y1~△Y4 are derived from Sphingomonas paucimobilis ATCC 31461, And △Y5~△Y7 are derived from Sphingomonas paucimobilis DSM 6314) inoculated on the slant of YM medium, cultured at 30°C for 72h;
[0066] 2. First-level seed culture: respectively insert slant seeds into 50ml first-level seed medium (filled in a 250ml Erlenmeyer flask), and culture at 30°C and 200rpm for 24 hours, which is the first-level seed liquid;
[0067] 3. Secondary seed culture: Inoculate the primary seed solution with 5% volume ratio into 100ml secondary seed culture medium (filled in a 500mL triangular flask), shake and cultivate at 30°C and 200rpm for 12h, which is the secondary seed solution. seed liquid;
[0068] 4. Fermentation: Put the secondary seed liqu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com