Sphingomonas alginate lyase gene ZH0-II as well as prokaryotic expression vector and application thereof
A technology of alginate lyase, ZH0-II, applied in the field of preparation of alginate lyase ZH0-II protein, can solve the problems of difficult separation of enzyme and degradation product, low enzyme production of wild bacteria, high production cost, etc. Achieving broad substrate specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Sphingomonas Sphingomonas Preparation and detection of sp. ZH0 genomic DNA
[0040] Sphingomonas used in the present invention Sphingomonas sp. ZH0 (ZH0) is the strain screened by our laboratory. Genomic DNA of Sphingomonas ZH0 was prepared by the extraction method of common bacterial genome. Use up the supernatant and collect the bacteria; add 100ul Solution I suspension bacteria, then add 30ul 10%SDS; 1ul 20mg / ml proteinase K, mix well, and incubate at 37°C for 1 hour; add 100ul 15mol / L NaCl, mix well; add 20ul CTAB / NaCl solution (CTAB 10%, NaCl 0.7mol / L), mix well, 65°C, 10 minutes; add an equal volume of phenol / chloroform / isoamyl alcohol 25:24:1) and mix well, centrifuge at 12000rpm for 5 minutes; Take the supernatant, add 2 times the volume of absolute ethanol, 0.1 times the volume of 3mol / L NaOAC, and place it at -20°C for 30 minutes; centrifuge at 12000rpm for 10 minutes; add 70% ethanol to the precipitate to wash; after the precipitate is drie...
Embodiment 2
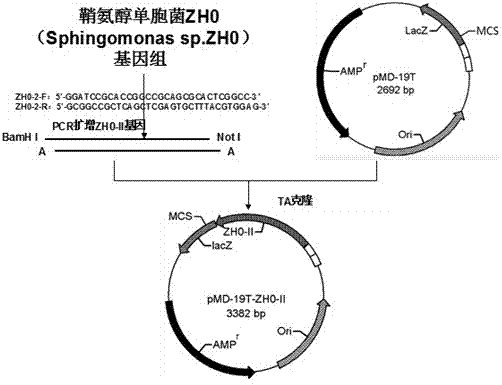
[0041] Example 2: Alginate Lyase Gene ZH0-II Amplification and TA cloning:
[0042] alginate lyase gene ZH0-II Amplification and TA cloning strategies such as figure 2 As shown, a pair of specific primers were designed according to the conserved sequence of the N-terminal and C-terminal of Sphingomonas alginate lyase, the sequence is as follows:
[0043] ZH0-2-F: GGATCCGCACCGGCCGCAGCGCACTCGGCC
[0044] ZH0-2-R: GCGGCCGCTCAGCTCGAGTGCTTTACGTGGAG
[0045] The primer at the 5' end has the GGATCC characteristic sequence, which forms a BamH I restriction site; the 3' end adds a GCGGCC characteristic sequence to form a Not I restriction site;
[0046] Add 10ng of Sphingomonas ZH0 genomic DNA as a template to the PCR reaction mixture, and add 50ng of specific primers ZH0-2-R and ZH0-2-F, 1.8uldNTP (10mM), 2.5ul of Pfu Reaction buffer and 0.15ul pfu (5U / ul) polymerase (Beijing Zhuangmeng International Biogene Technology Co., Ltd.), add double distilled water to make the final vo...
Embodiment 3
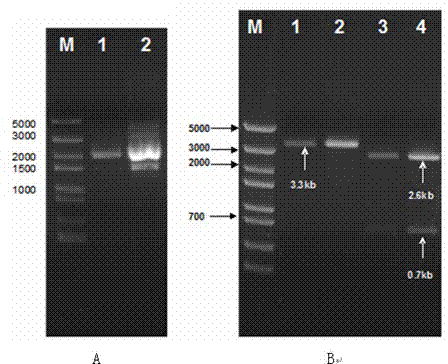
[0047] Example 3: Prokaryotic expression vector pGEX-4T-1- ZH0-II build
[0048] pGEX-4T-1- ZH0-II A build strategy such as Figure 4 As shown, the purified prokaryotic expression vectors pGEX-4T-1 (purchased from GE Healthcare) and pMD19- ZH0-II , separated the cleaved vector and insert fragments by agarose gel electrophoresis, and recovered the vector fragments pGEX-4T-1 (4.9kb) and pMD19- ZH0-II produced by cutting ZH0-II The DNA fragment (0.7kb) of the gene, and then use the ligase kit of TaKaRa to connect the pGEX-4T-1 vector fragment and ZH0-II The DNA fragment of the gene produces the prokaryotic expression vector pGEX-4T-1- ZH0-II , converting the high efficiency (10 8 ) Escherichia coli competent cells (JM109, Beijing Zhuangmeng International Biogene Technology Co., Ltd.), spread the transformed Escherichia coli on the plate of ampicillin (Amp, 100ug / ml), and cultivate overnight at 37°C , screen Amp-resistant recombinant colonies, and extract plasmids from Amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com