Method for constructing genetically engineered Escherichia coli using xylose metabolism to produce succinate
A technology of genetically engineering bacteria and producing succinic acid, applied in the field of bioengineering, can solve the problems of wasting resources, polluting the environment, etc., and achieve the effect of efficient utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
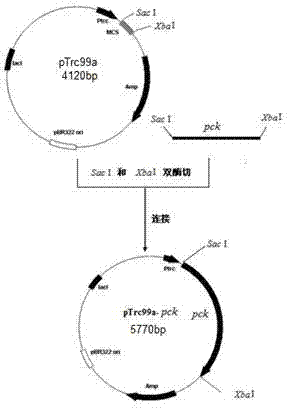
[0054] This example illustrates the construction of an expression plasmid that overexpresses phosphoenolpyruvate carboxykinase, restores the ability of the recombinant strain to metabolize xylose under anaerobic conditions, and obtains the strain Escherichia coli BA204.
[0055] 1. Lack of lactate dehydrogenase gene ( wxya ), pyruvate formate lyase gene ( PPML ) Active E. coli The NZN111 strain was the starting strain, and the phosphoenolpyruvate carboxylase (PPC) gene was knocked out to obtain simultaneous deficiency wxya , PPML and PPC competent strains.
[0056] Knockout of the phosphoenolpyruvate carboxylase (PPC) gene using homologous recombination technology: using the apramycin resistance gene with FRT sites on both sides as a template, using a high-fidelity PCR amplification system, and designing Amplification primers with PPC homologous fragments at both ends successfully amplified linear DNA homologous fragments; a plasmid capable of inducing the expressi...
Embodiment 2
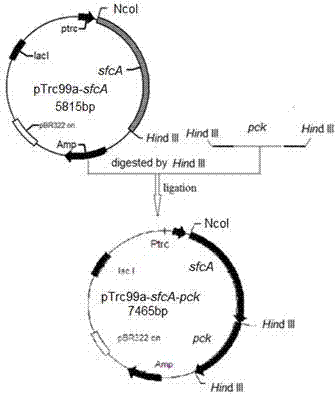
[0064] This example illustrates the construction of an expression plasmid co-expressing phosphoenolpyruvate carboxykinase and malic enzyme, restoring the ability of the recombinant strain to metabolize xylose under anaerobic conditions, and obtaining the strain Escherichia coli BA205.
[0065] 1. Lack of lactate dehydrogenase gene ( wxya ), pyruvate formate lyase gene ( PPML ) Active E. coli The NZN111 strain was the starting strain, and the phosphoenolpyruvate carboxylase (PPC) gene was knocked out to obtain simultaneous deficiency wxya , PPML And the competent bacterial strain of PPC (same as embodiment 1).
[0066] 2. Construction of an expression plasmid co-expressing phosphoenolpyruvate carboxykinase and malic enzyme, the process comprising:
[0067] (1) Both the upstream and downstream of the synthesis have Hin primers for the dIII restriction site,
[0068]Upstream primer: 5'- CCCAAGCTTATGAACTCAGTTGATTTGACCG -3';
[0069] Downstream primer: 5'-CCCAAGCTTG...
Embodiment 3
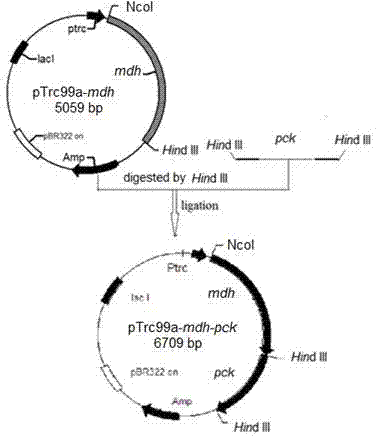
[0073] This example illustrates the construction of an expression plasmid co-expressing phosphoenolpyruvate carboxykinase and malate dehydrogenase, restoring the ability of the recombinant strain to metabolize xylose under anaerobic conditions, and obtaining the strain Escherichia coli BA206.
[0074] 1. Lack of lactate dehydrogenase gene ( wxya ), pyruvate formate lyase gene ( PPML ) Active E. coli The NZN111 strain was the starting strain, and the phosphoenolpyruvate carboxylase (PPC) gene was knocked out to obtain simultaneous deficiency wxya , PPML And the competent bacterial strain of PPC (same as embodiment 1).
[0075] 2. Construction of an expression plasmid co-expressing phosphoenolpyruvate carboxykinase and malate dehydrogenase, the process comprising:
[0076] (1) Both the upstream and downstream of the synthesis have Hin Primers for the dIII restriction site,
[0077] Upstream primer: 5'- CCCAAGCTTATGAACTCAGTTGATTTGACCG -3';
[0078] Downstream pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com