Same sequence primer transpositional nucleic acid amplification technology
A technology of nucleic acid and technology, applied in the field of nucleic acid amplification, which can solve the problems of delay, inability to eliminate, and limited degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
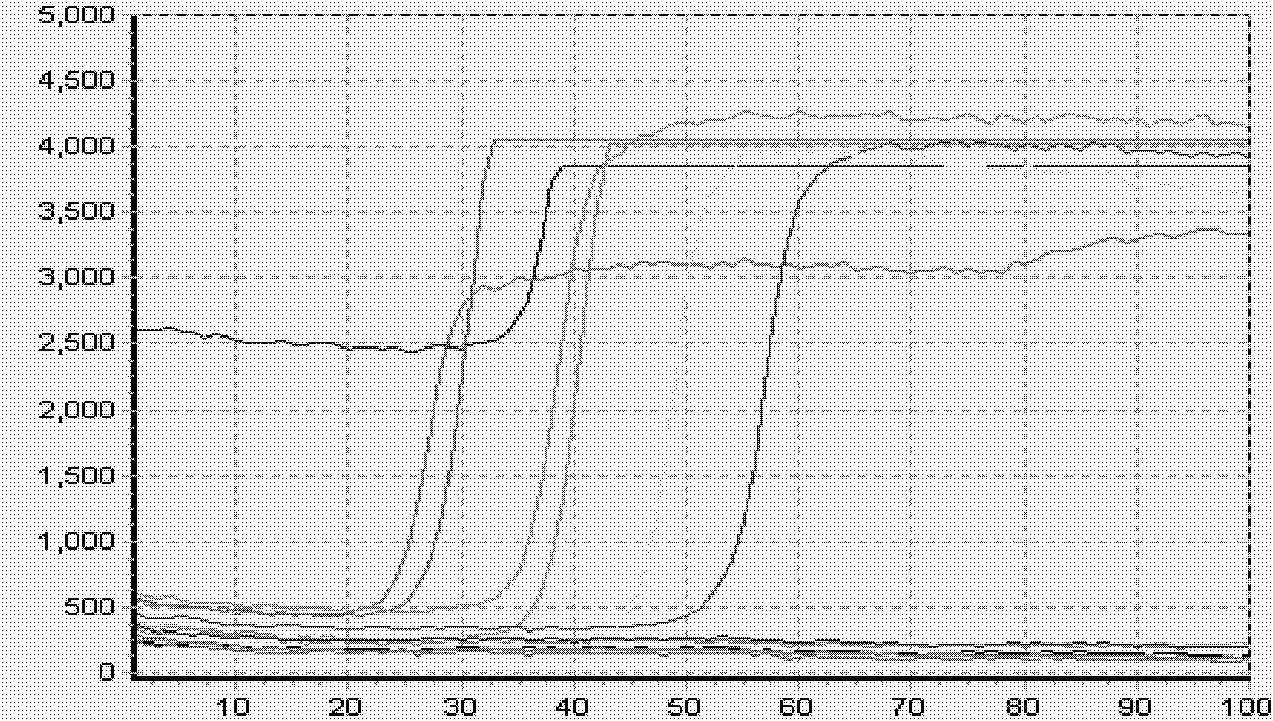
[0093] Example 1: Human hepatitis B virus homogeneous primer displacement fluorescent PCR detection:
[0094] Hepatitis B virus (referred to as hepatitis B) is a worldwide infectious disease caused by hepatitis B virus (Hepatitis B virus, HBV). The infection rate of hepatitis B in our country is very high, which greatly endangers people's health. At present, the detection methods of hepatitis B mainly include enzyme immunoassay, radioimmunoassay, chemiluminescence, immunofluorescence, nucleic acid amplification (PCR) fluorescence quantitative method and so on. Enzyme immunoassay is widely used, but real-time fluorescent PCR analysis can accurately detect the virus gene of hepatitis B patients, and plays an irreplaceable important role in judging the virus replication level of infected patients, monitoring the infectivity of the disease and the efficacy of antiviral drugs.
[0095] Hepatitis B virus (HBV) is a partially double-stranded DNA virus. It mainly has three type-speci...
example 2
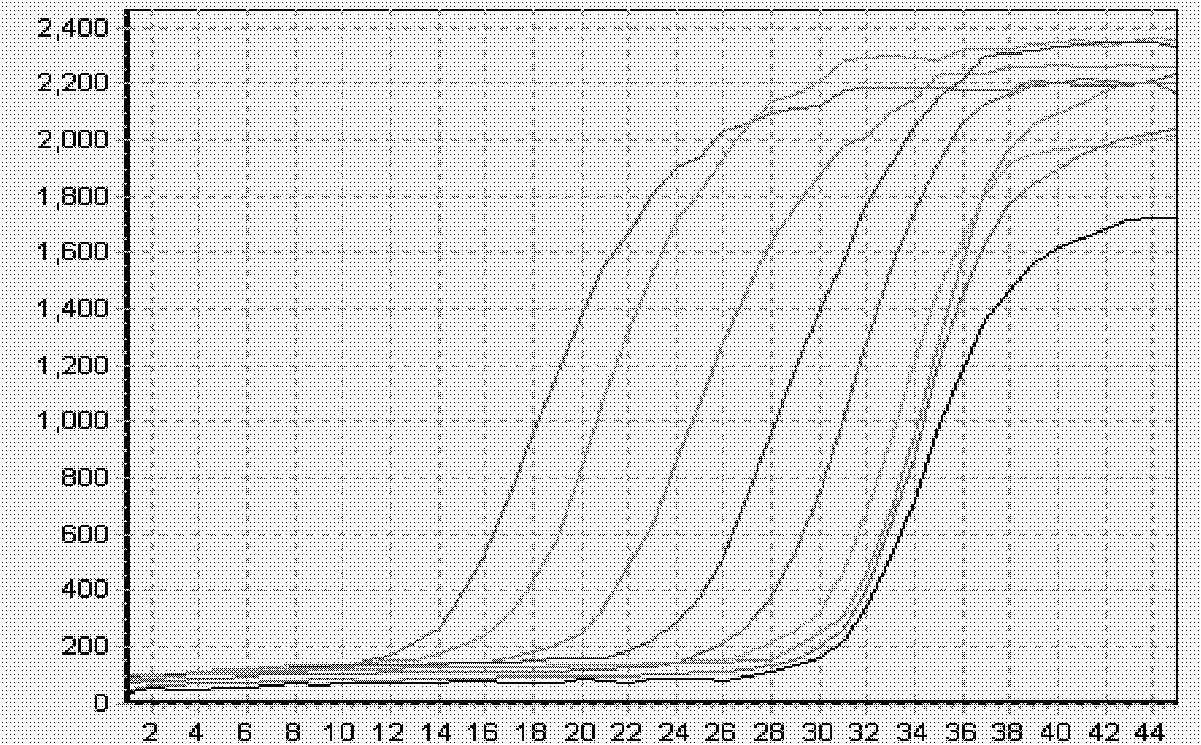
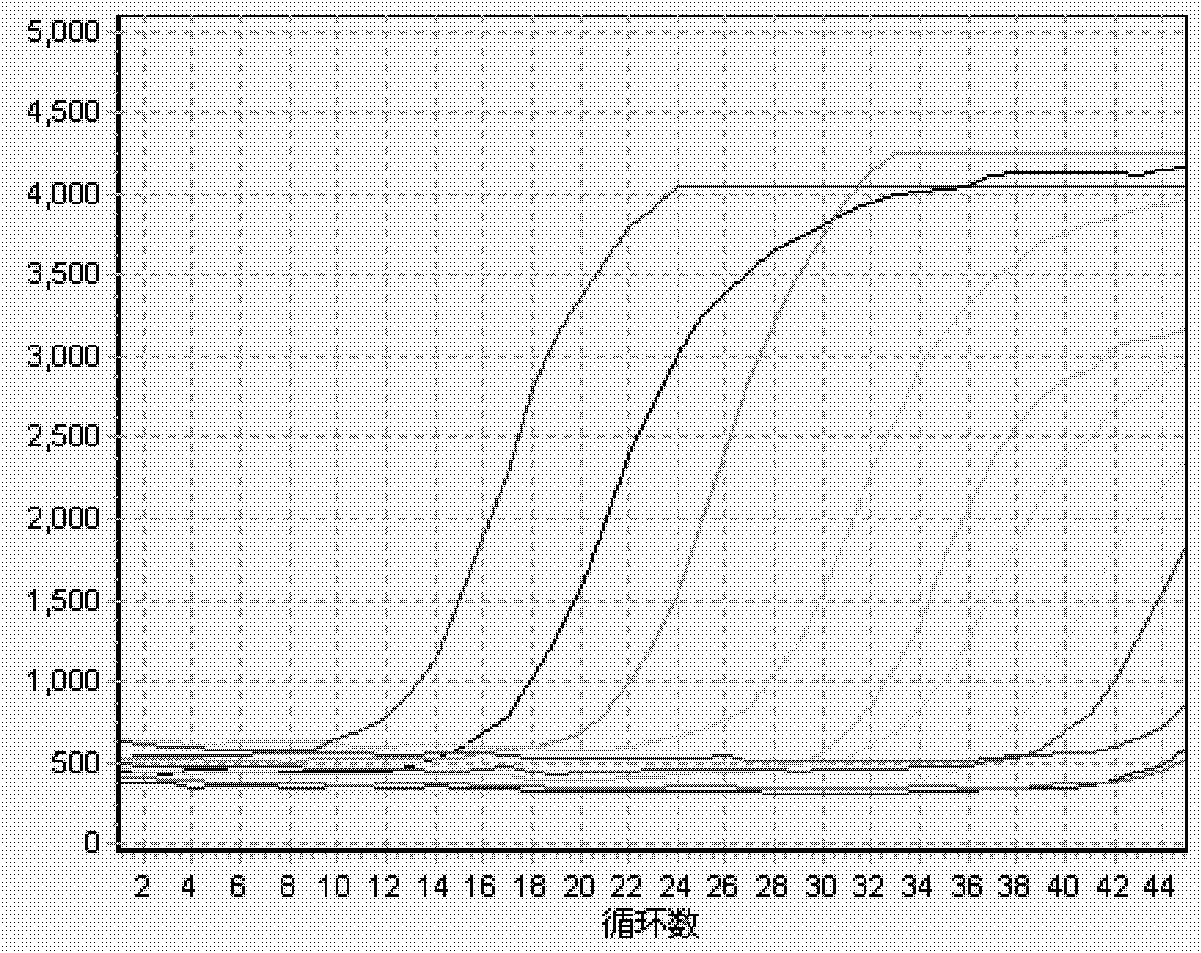
[0126] Example 2: Double fluorescent PCR detection of human enterovirus pathogenic strains:
[0127] In recent years, hand, foot and mouth disease has become prevalent among young children in my country, and the case fatality rate is high. The nucleic acid real-time fluorescent PCR detection of its pathogenic enterovirus (EnteroVirus, EV) has become a key means to control its epidemic. Enterovirus EV is initially divided into more than 60 different serotypes, including enterovirus 68-71. Based on its nucleic acid sequence classification, human EVs are further divided into five categories: A, B, C, D and PolioVirus, among which the main pathogenic strains Coxsackie A16 (CA16) and EnteroVirus 71 (EV71) are classified as human enterovirus A. The EV gene of enterovirus is highly variable, only the 5'UTR is conserved, and there are three homologous conserved regions of all strains. The published EV universal primers all select this conserved region, so they misdetect non-pathogenic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com