Expression of recombined human protein disulphide isomerase (hPDI491) with Pichia pastoris in secretion manner
A disulfide bond isomerase and protein technology, applied in the direction of isomerase, recombinant DNA technology, application, etc., to achieve the effect of high expression, high activity, and reducing the formation of protein inclusion bodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Design and construction of recombinant plasmids, preparation of r-hPDI 491 protein
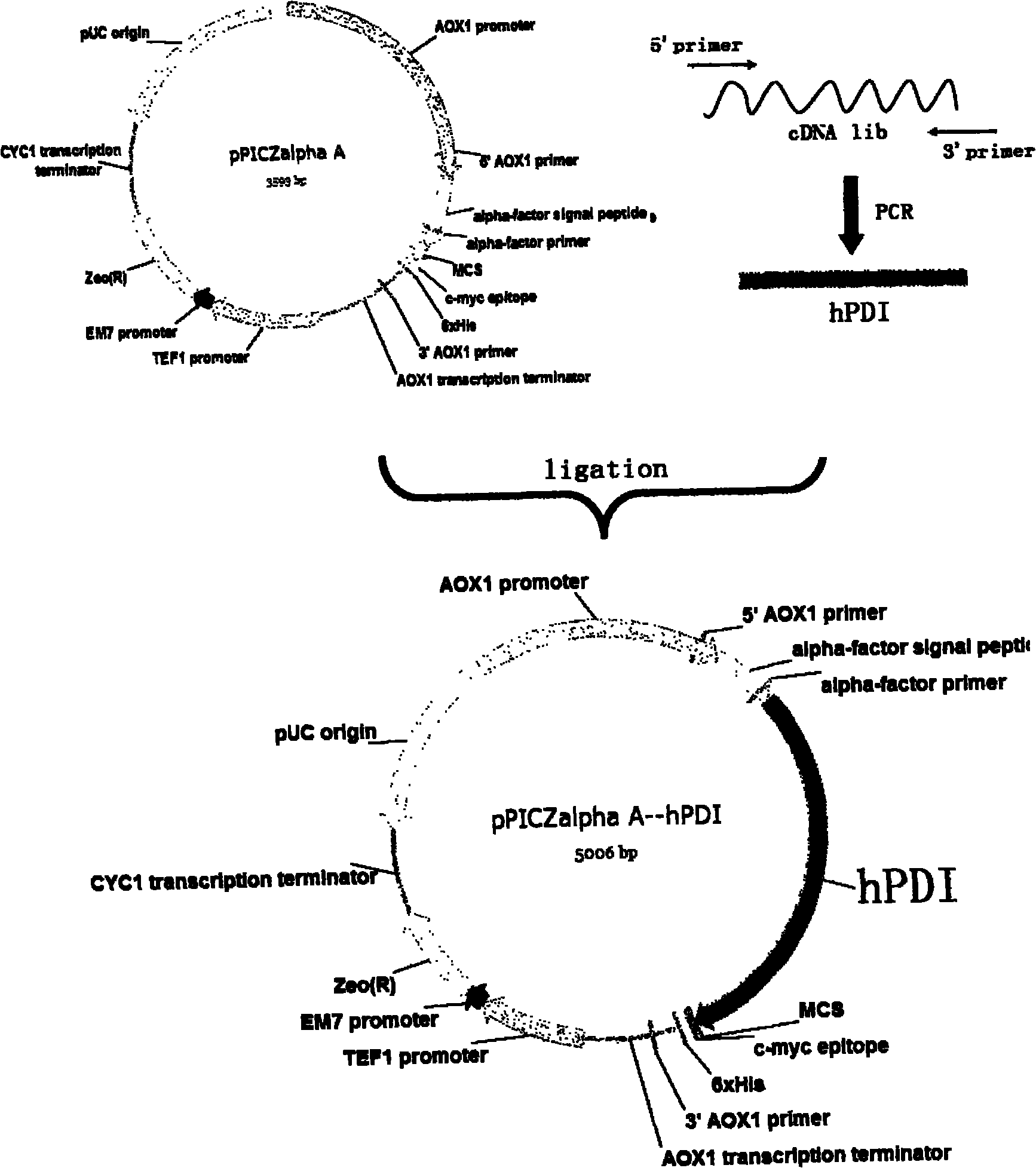
[0027] (1) Clone hPDI 491 cDNA gene, construction of expression plasmid pPICZα-hPDI 491
[0028] According to hPDI 491 Gene sequence, design primer sequence. The target gene was amplified from the human liver cDNA library by PCR method. The amplified product was excised from the restriction endonuclease site, and recombined with the expression plasmid pPICZα of Pichia pastoris to construct the expression plasmid pPICZα-hPDI 491 , Transform NovaBlue host bacteria. Extract the plasmid, and then analyze the corresponding restriction endonuclease site to screen the plasmid with characteristic fragments, and then use nucleotide sequence analysis to confirm the correct position and sequence of gene recombination, and obtain positive clones.
[0029] The restriction endonuclease used in the present invention was purchased from Takara Company, the methanolotrophic Pichia strai...
Embodiment 2
[0049] Example 2: r-hPDI 491 Protein Bioactivity Assay
[0050] r-hPDI 491 Protein-catalyzed granule monolineage colony-stimulating factor (GM-CSF) folding assay. After the GM-CSF inclusion body protein extracted with urea was purified by SephacrylS-200, it was added to the TKM buffer at a concentration of 1 mg / mL, and then 4 mg / mL r-hPDI was added 491 and 10 5 mol / L DTT, acted at 25°C for 10 hours, measured the activity without adding r-hPDI 491 The GM-CSF activity in the above system was used as a control, and the molecular state was detected by gel exclusion HPLC. At the concentration of GM-CSF 1mg / mL with Cu 2- Oxidative refolding, the specific activity of GM-CSF was 4.2×10 6 u / mg, monomer peak area accounted for 62%, r-hPDI 491 Under protein catalysis, the specific activity can be increased to 8.5×10 6 u / mg, the monomer peak area increased to 92%.
[0051] DNA and protein molecular sequences
[0052] SEQ-1 sequence description:
[0053] (1) Sequence description...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com