Specific sequence characterized amplified region (SCAR) marker for Trialeurodes vaporariorum, specific primers, and quick molecular identification method
A technology for labeling primers and whitefly, applied in the field of molecular biology, can solve the problem of not being able to identify molecular markers at the same time, and achieve the effects of rapid and accurate identification, cheap identification and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Single Whitefly Pest DNA Extraction
[0050] Genomic DNA was extracted using Genome Extraction Kit from Biotek Technology Co., Ltd., and the specific operations were as follows: (1) Place the single-headed whitefly on the Parafilm membrane dripped with 10 μL of lysate, and use the bottom of a 0.2 mL PCR tube as a uniform sample. Grind the homogenizer thoroughly, wash the homogenizer twice with another 20 μL of lysate, combine and mix; (2) Transfer to a 1.5 mL centrifuge tube with a micropipette, add 2 μL proteinase K, mix well, and place in a water bath at 55 °C for 4 hours Or overnight; (3) Centrifuge for a few seconds, add 2.5 μL of RNase, and place in a 37°C oven for 15-30 minutes; (4) Take out the above sample, let it cool to room temperature, add 25 μL of protein precipitation solution, and shake at high speed on a vortex shaker for 25 seconds; (5) Ice bath for 5 minutes to precipitate protein; centrifuge at room temperature, 13000r / min, centrifuge for 5 ...
Embodiment 2
[0051] Embodiment 2RAPD amplification
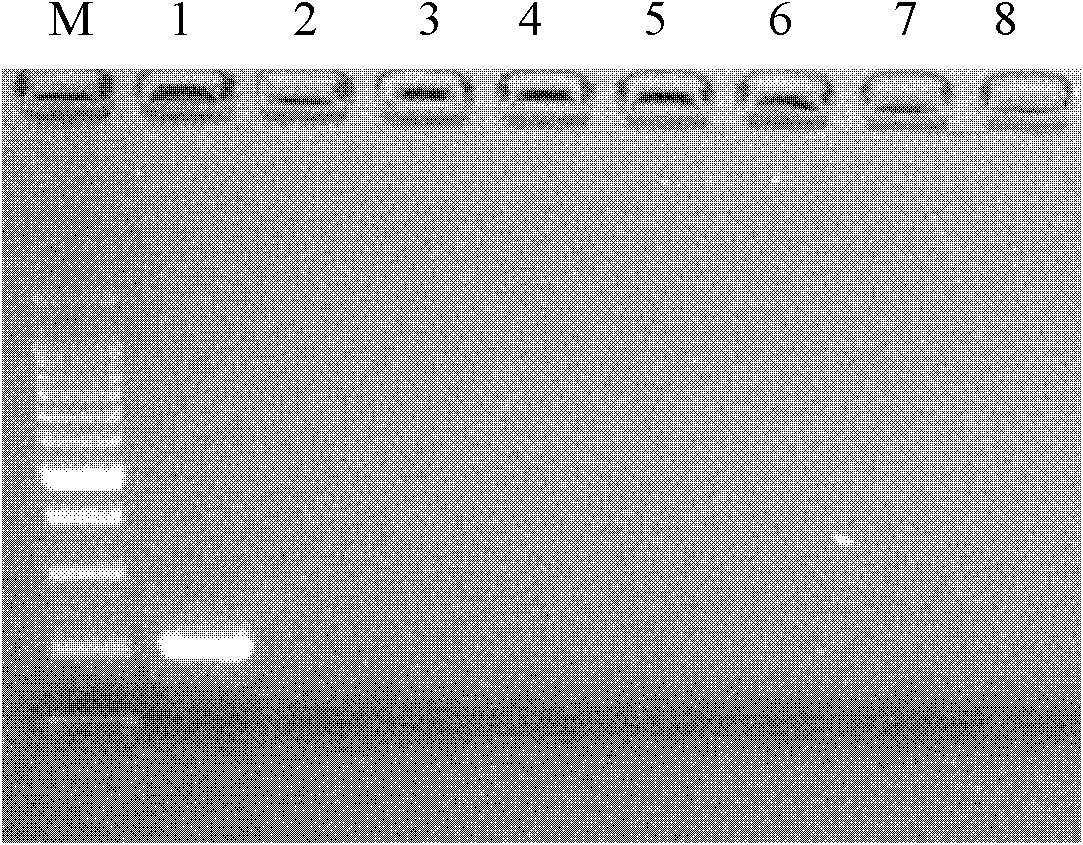
[0052] Using the genomic DNA of the test sample in the above materials and methods as a template, 60 random primers were used for random amplification, and the screened specific bands and corresponding amplification primers were recorded, and these bands were labeled. The total volume of amplification is 20 μL, including 10 μL 2×Mix, 1 μL DNA template, 3 μL RAPD primer (10 mM) and 6 μL dd H 2 O. The PCR reaction program was as follows: first, 94°C pre-denaturation for 4 min, followed by 40 cycles, including 94°C for 1 min, 37°C for 50 s, 72°C for 1 min and 35 s, and finally 72°C for 10 min. Take 6 μL of the PCR amplification product of each sample and load it on electrophoresis.
[0053] In RAPD random amplification, it was found that when OPA-17 primer was amplified, it showed strong species specificity for greenhouse whitefly, and a 620bp DNA band appeared in greenhouse whitefly, and the specific band was very bright , and appeared ...
Embodiment 3
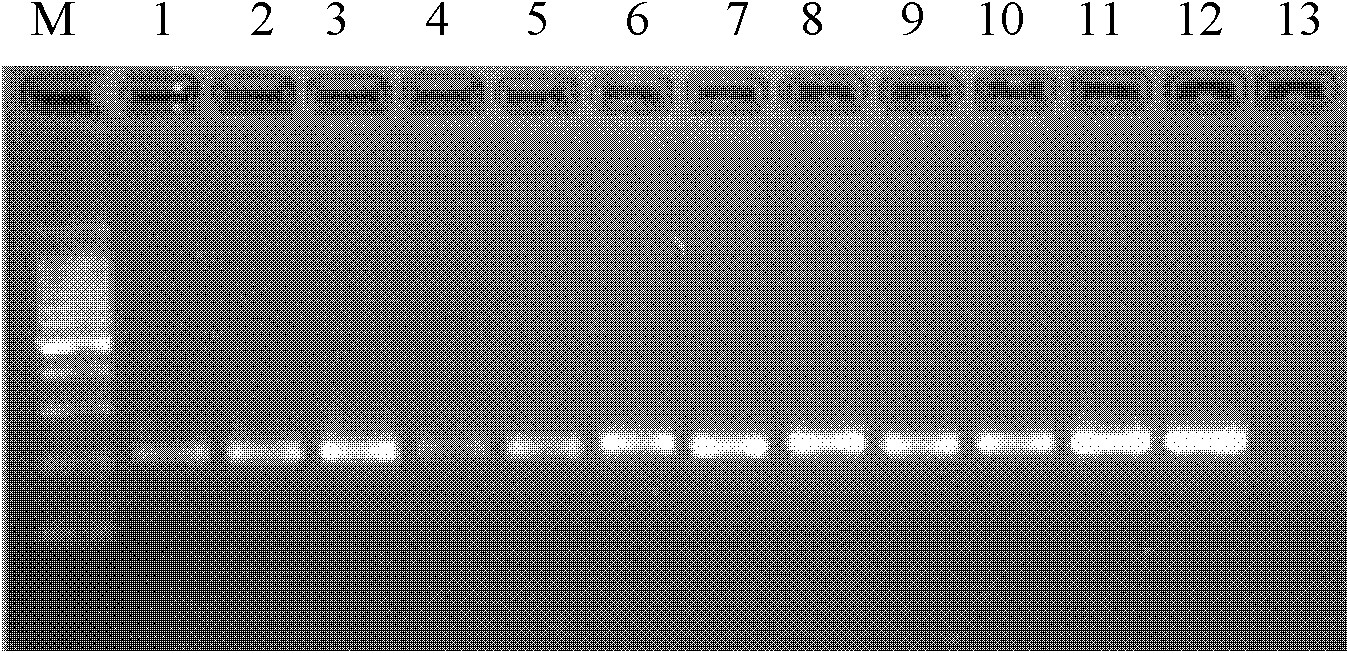
[0054] Example 3 Recovery Cloning of Specific Amplified Products, SCAR Primer Design and PCR Amplification
[0055] Mark the specific bands found in the greenhouse whitefly samples in the RAPD amplification results. The specific band was recovered on agarose gel electrophoresis, purified using a DNA purification kit (Biteke, Beijing), and the purified fragment was connected to the pEASY-T1 vector (Quanjin, Beijing) and transformed into TOP10 On the competent cells, culture them upside down at 37°C overnight, pick white positive clones, add them to LB liquid medium for shaking culture at 37°C overnight, and finally use the plasmid extraction kit from Beijing Biotek Biotechnology Co., Ltd. to extract the plasmids, positive plasmids After being amplified and verified by the forward and reverse primers of the universal primer M13, it was sent to Beijing Jiaoqingke Bioengineering Co., Ltd. for sequencing.
[0056] The sequencing result is shown in SEQ ID NO.1:
[0057] gaccgcttgt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com