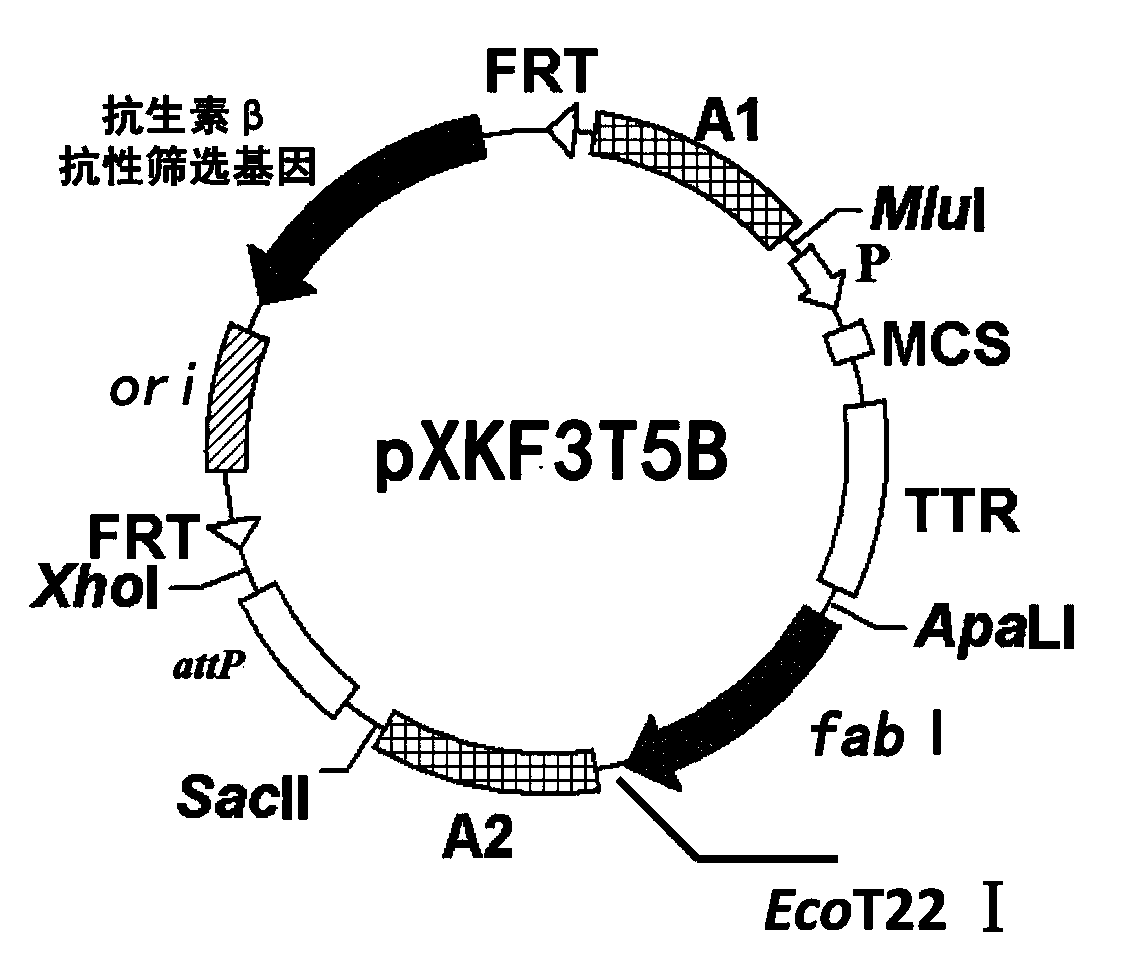
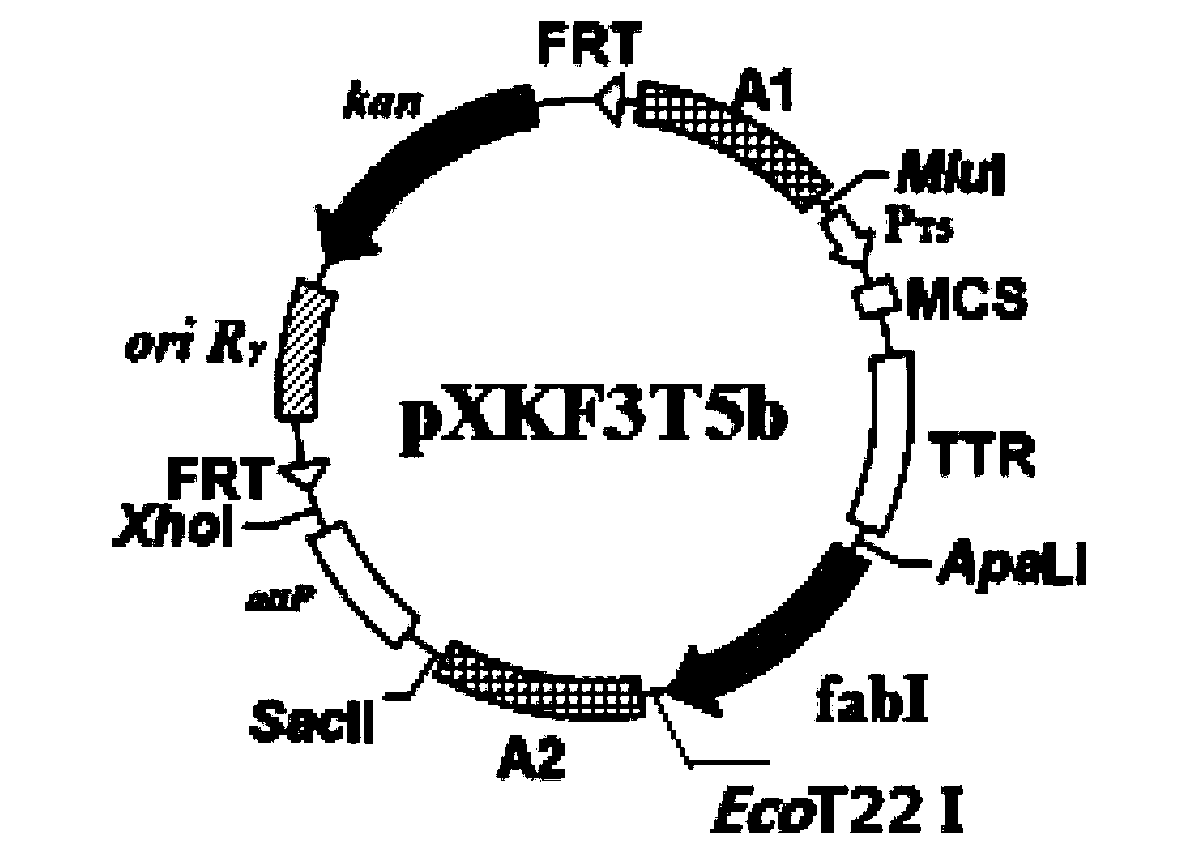
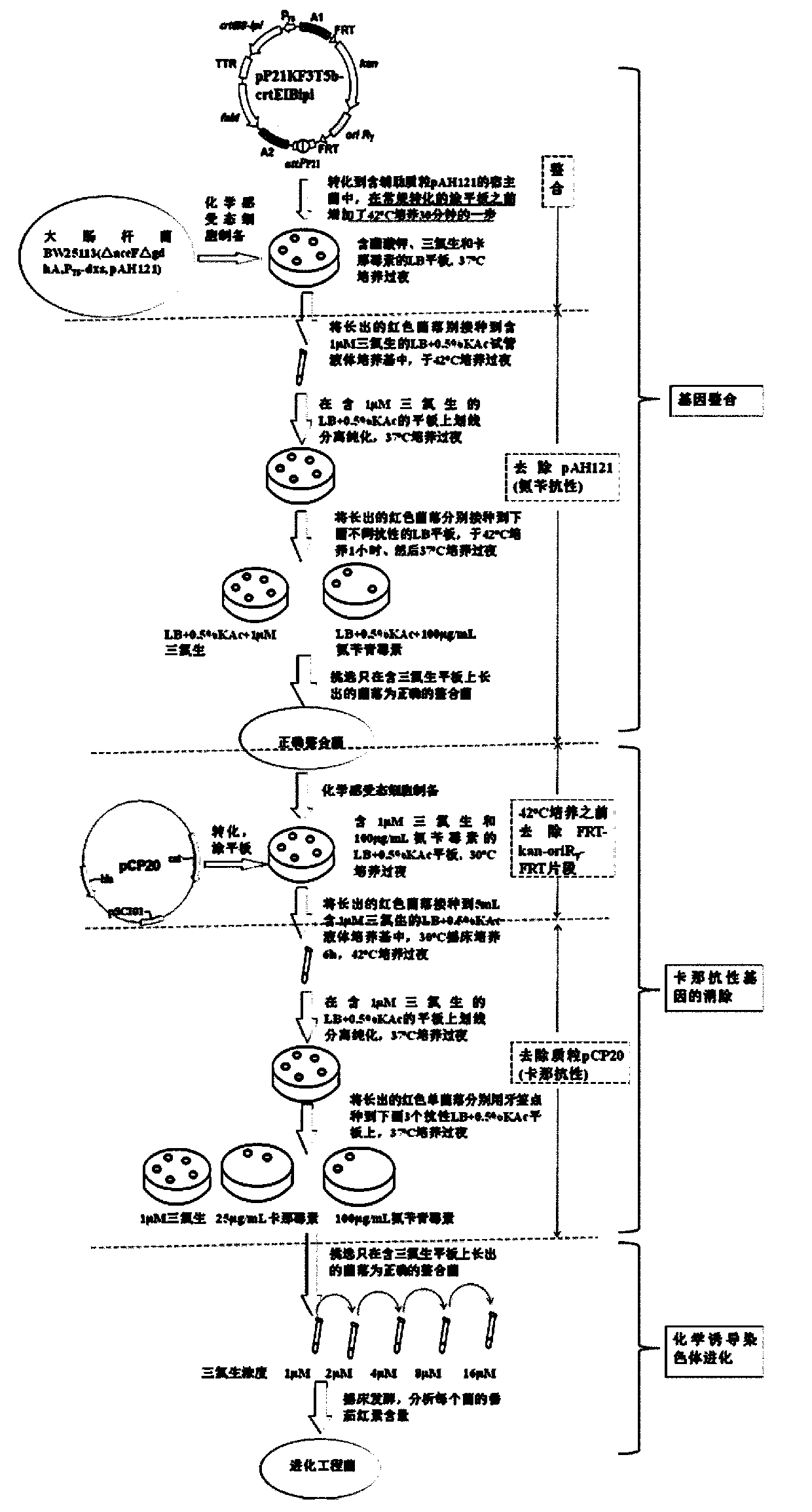
Construction method of gene engineering strain without plasmid and antibiotic resistance screening marker
A technology of resistance screening markers and genetically engineered bacteria, applied in the field of bioengineering, can solve the problems of antibiotic resistance, complexity, and low integration efficiency, and achieve the effects of stable production performance and high genetic stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Construction of Lycopene-producing Engineering Bacteria Using Escherichia coli BW25113 as Starting Bacteria
[0052] 1) Transform the helper plasmid pAH121 expressing integrase (a gift from Professor Wanner of Purdue University, see Haldimann, A., Wanner, B.L. Journal of Bacteriology.2001, 183: 6384-6393) into E. coli E. coli BW25113 to obtain Escherichia coli E. coli BW25113 (pAH121).
[0053] 2) Hind III digestion of pB-crtEIBipi (gifted by Professor Liu Jianzhong of Sun Yat-Sen University, expression vector containing lycopene synthesis gene) and filled in, then digested with EcoR I, gel recovery of 4329bp lycopene synthesis gene crtEIBipi fragment , connected to the pP21KF3T5b vector (SEQ ID NO: 2) digested with EcoR I / BamH III to obtain pP21KF3T5b-crtEIBipi.
[0054] 3) Gene integration: transform pP21KF3T5b-crtEIBipi into Escherichia coli E.coli BW25113(pAH121): add 5 μl of pP21KF3T5b-crtEIBipi to 100 μl of E.coli BW25113(pAH121) competent cells, and place on ice...
Embodiment 2
[0064] Escherichia coli BW25113 (ΔgdhAΔaceF, P T5 -dxs) is the construction of the lycopene-producing engineering bacterium of the starting bacterium
[0065] Change Escherichia coli E.coli BW25113 in Example 1 into Escherichia coli E.coli BW25113 (ΔgdhAΔaceF, P T5 -dxs) (gifted by Professor Liu Jianzhong of Sun Yat-sen University), the lycopene synthesis gene crtEIBipi was connected to the chromosomal evolution plasmid pHKKF3T5b to obtain pHKKF3T5b-crtEIBipi, and the helper plasmid for expressing integrase was pAH69, and the other steps were the same as in Example 1. Finally, the chromosomally evolved E.coli CIChE-crtEIBipi tolerant to 8 μM triclosan was obtained, and the shaker fermentation could produce 30.5 mg / g dry lycopene.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com