EMA-PCR (Ethidium Monoazide-Polymerase Chain Reaction) detection method of viable pathogen in farm environment
A detection method and farm technology are applied in the field of EMA-PCR detection of live bacteria, which can solve the problems of false positive detection results, inability to distinguish dead bacteria from live bacteria, and interference with the authenticity of detection, achieving simple operation and suitable for promotion. , the effect of short time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] This embodiment mainly consists of the following steps:
[0039] (1) The bacterial liquid is obtained after the sample is pretreated;
[0040] (a1) Take 1.5g wet weight stool sample;
[0041] (a2) Add 30ml of sterile PBS buffer, vortex to suspend, centrifuge at 500×g for 4min, and take the supernatant;
[0042] (a3) Repeat step (a2) for the precipitate formed after centrifugation; centrifuge the obtained supernatant at 9000×g for 5 minutes at high speed;
[0043] (a4) Collect the precipitate, suspend it with 30ml of 0.1M PBS buffer, and centrifuge at 9000×g for 5min;
[0044] (a5) After repeating step (a4), the bacterial cells were obtained, and the bacterial cells were suspended in 3ml of TE buffer solution, shaken and mixed evenly, and the bacterial liquid was prepared.
[0045] (2) Add 100ug of ethidium azide bromide to 1ml of the bacterial solution, place it in the dark for 5 minutes, and treat it with a 650W halogen light source for 1 minute. In this embodiment...
Embodiment 2
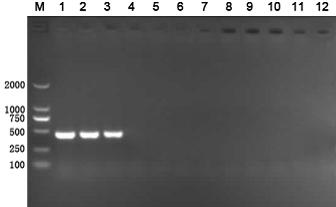
[0058] The difference between this embodiment and Example 1 is that the bacterial species detected are different. The bacterial species detected in this embodiment is enterohaemorrhagic Escherichia coli O157:H7. Taking the rfbE specific amplification primer as an example, the primer sequence is: F : 5'-AGATTGCGCTGAAGCCTTTG-3'; R: 5'-ATTGGCATCGTGTGGACAG-3', the size of the amplified target band is 495bp, and the detection result is as follows Figure 5 .
[0059] Figure 5 1 to 3 are the detection results of enterohemorrhagic Escherichia coli O157:H7, and 4 to 12 are the detection results of the control strain.
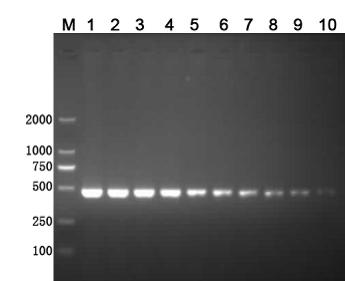
[0060] The control experiment of the present embodiment is: the EMA-PCR amplification of the mixed bacterial suspension of different viable cell ratios, and detection result is as follows Figure 6 .
Embodiment 3
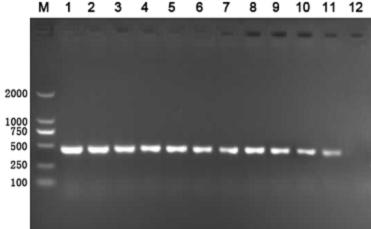
[0062] The difference between this embodiment and Example 1 is that the strains detected are different. The strains detected in this embodiment are Staphylococcus aureus. Taking the nuc-specific amplification primer as an example, the primer sequence is: F:5'- TTAGCCAAGCCTTGACGAAC-3′; R: 5′-AGGGCAATACGCAAAGAGGT-3′, the size of the amplified target band is 480bp, and the detection result is as follows Figure 7 .
[0063] Figure 7 1 to 4 are the test results of Staphylococcus aureus, and 5 to 12 are the test results of the control strain.
[0064] The control experiment of the present embodiment is: the EMA-PCR amplification of the mixed bacterial suspension of different living cell ratio, detection result is as follows Figure 8 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com