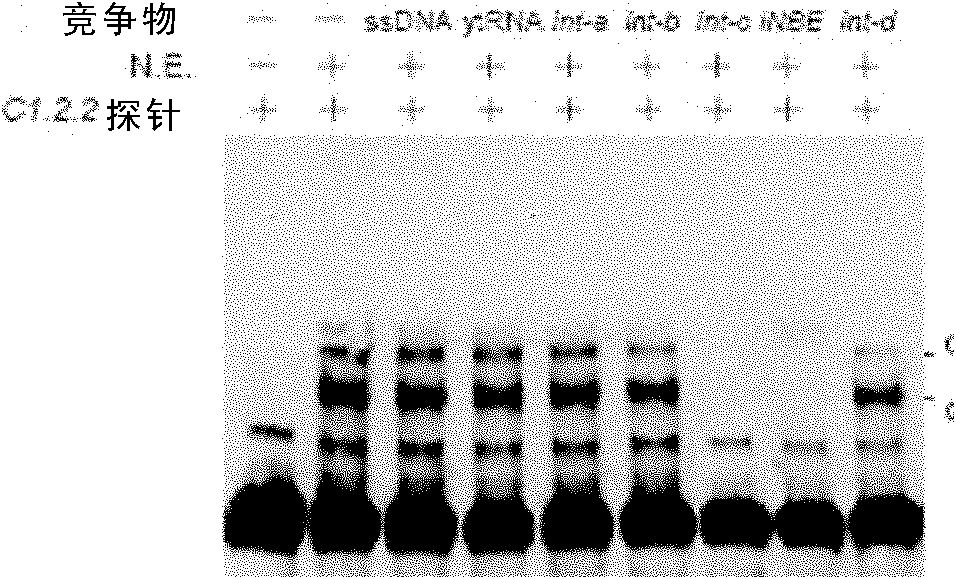DNA sequence regulating eukaryotic gene transcription, and its binding proteins
A gene and actin technology, applied in the field of DNA sequences regulating eukaryotic gene transcription and their binding proteins, can solve problems such as unclear molecular mechanisms of acting factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
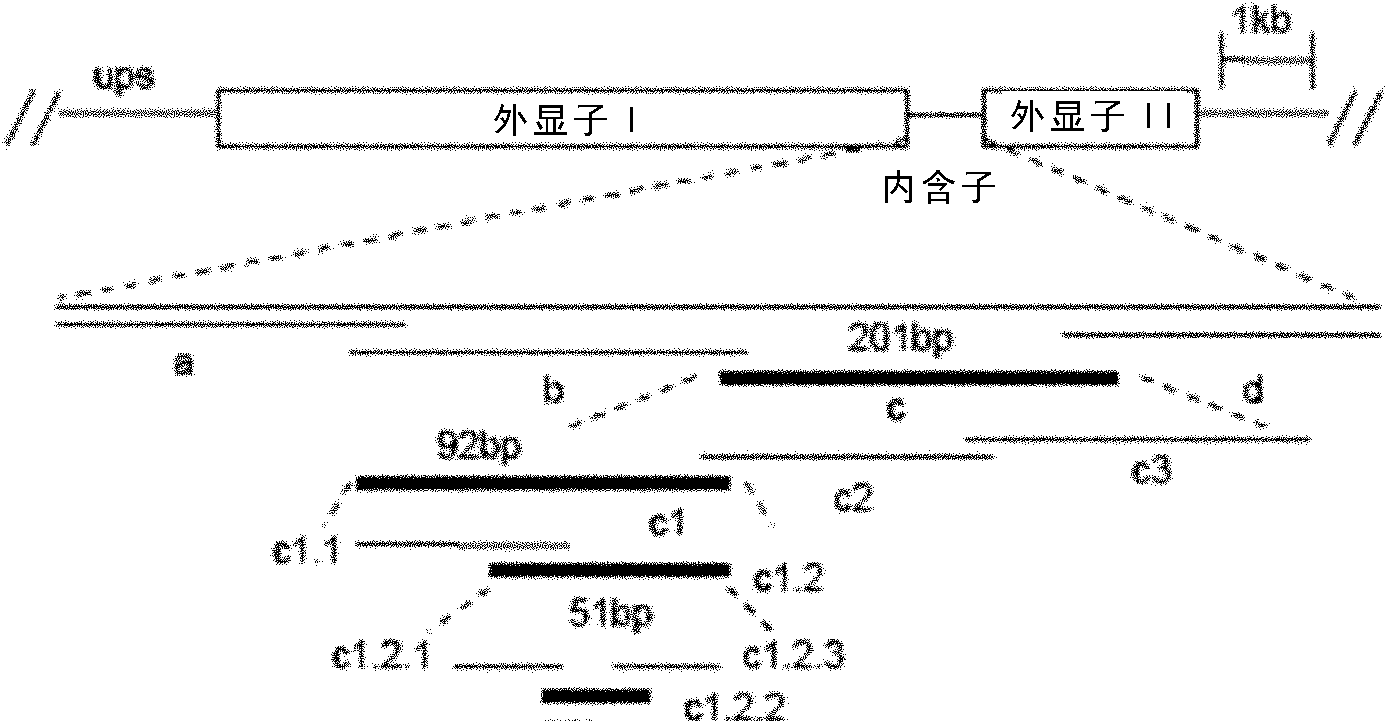
[0085] Example 1, Amplification of Plasmodium falciparum var gene intron sequence and probe preparation
[0086] (1) Intron sequence amplification of var gene
[0087] In order to prepare EMSA probes, it is first necessary to amplify the var gene intron sequence from the genome of Plasmodium falciparum 3D7 strain (see Chinese patent application 200610118947.8), clone it into a vector and perform sequencing. Since the AT content in the non-coding region of Plasmodium falciparum can be as high as more than 90%, ordinary Taq enzymes cannot effectively amplify these sequences, and it is difficult to ensure the correctness of the sequences. In this example, in order to balance amplification efficiency and sequence fidelity, the inventors all used Toyobo's KOD-Plus high-fidelity enzyme. The specific reaction preparation and amplification procedures are as follows:
[0088] 10x KOD-Plus reaction buffer: 5 μl;
[0089] MgCl (2mM): 5μl;
[0090] dNTP (2mM): 5μl;
[0091] Upstream ...
Embodiment 2
[0108] Example 2, Preliminary Extraction of Total Nucleoprotein of Plasmodium falciparum Hainan Strain
[0109]1. Get 100ml of Plasmodium falciparum 3D7 strain culture cultured in vitro, the parasite rate is about 10% (the number of erythrocytes infected by Plasmodium per 100 erythrocytes), 2000rpm, 5min to remove the supernatant.
[0110] 2. Add 20ml of 0.15% (w / v) saponin / PBS solution, mix well, 4°C, 30min.
[0111] 3. 8000rpm, 4°C, 10min, remove the supernatant.
[0112] 4. Wash 3 times with 20ml of cold 1xPBS solution, 4°C, 8000rpm, 10 minutes each time.
[0113] 5. Add 1.5ml complete cell lysate (20mM Hepes pH7.9, 10mM KCl, 1mM EDTA, 1mM EGTA, 1mM DTT, 1× protease inhibitor mixture, 0.65% (v / v) NP-40), mix well, Shake for 10 seconds and place in an ice-water bath for 10 minutes.
[0114] 6. 8000rpm, 5min, carefully draw the supernatant, which is the Plasmodium cytoplasmic protein, and store it at -80°C after aliquoting.
[0115] 7. Repeat 2 times to fully wash the nuc...
Embodiment 3
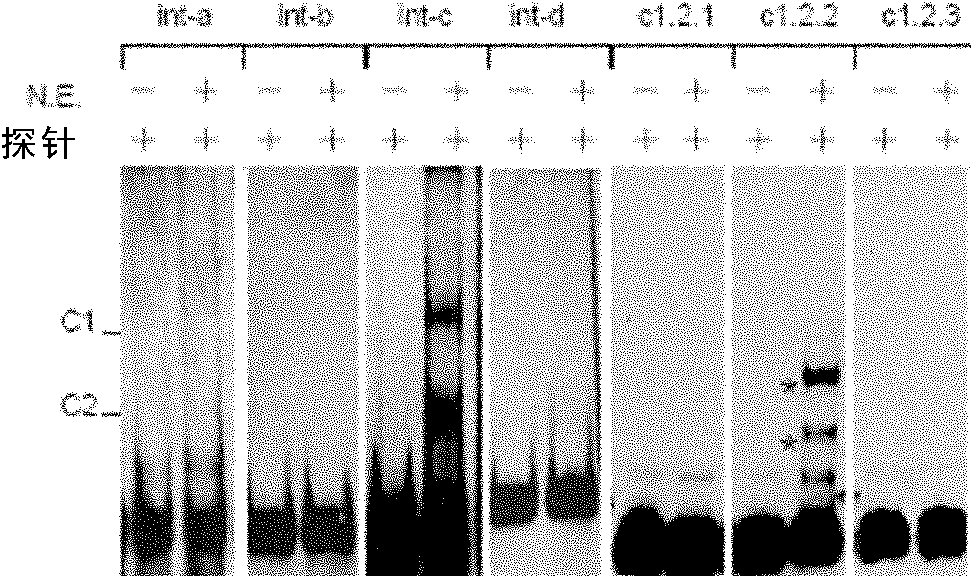
[0120] Embodiment 3, EMSA experiment identification intron-nucleoprotein complex
[0121] In this example, the inventor mainly used Pierce's LightShift Chemiluminescent EMSA kit to identify nucleoproteins (see the Pierce 20418 product manual for specific methods). Its main steps are as follows:
[0122] A. Preliminary experiment, initially establish the EMSA system, and explore the optimal amount of probe;
[0123] B. EMSA analysis
[0124] (1) Prepare the corresponding non-denaturing PAGE gel according to the size of the probe.
[0125] (2) Use 0.5×TBE as electrophoresis buffer, 4°C, 100V, pre-electrophoresis for 1 hour.
[0126] (3) According to different experimental purposes, design the corresponding EMSA reaction system (taking 20 μl system as an example), and each system is shown in Table 1.
[0127] Table 1
[0128]
[0129]
[0130] *In the table, the volume less than 20μl is supplemented with water.
[0131] **The preparation of the binding buffer (EMSA bu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com