Pichia stipitis genome-scale metabolic network model construction and analysis method
A metabolic network and genome technology, applied in educational tools, instruments, teaching aids, etc., can solve the problem of low substrate absorption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
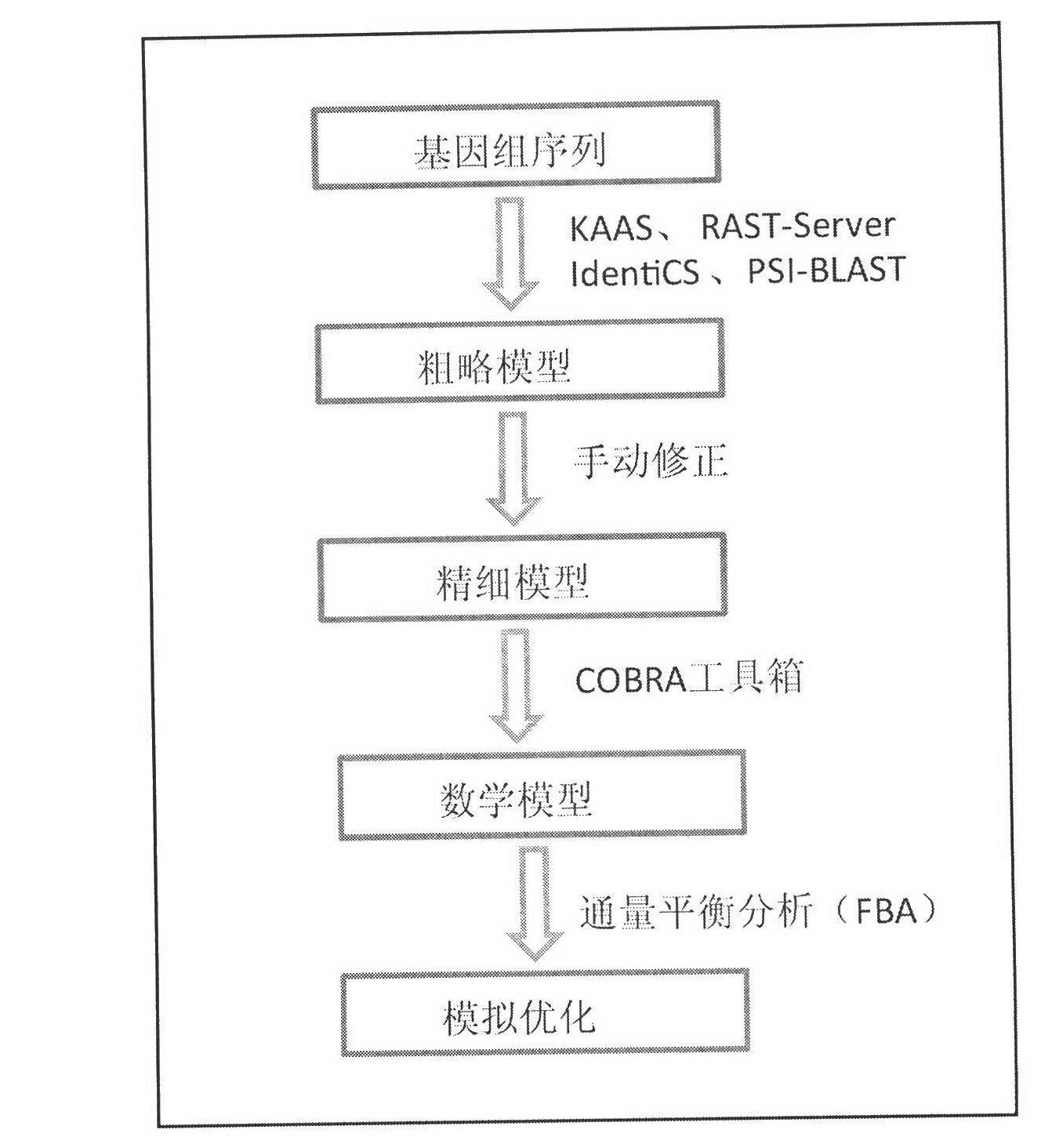
[0017] The construction process of the genome-scale metabolic network model of Pichia stipitis:
[0018] 1) Download the latest Scheffersomyces stipitis CBS6054 genome sequence from the NCBI Entre Gene database, and use the following three methods to annotate the genome. ①Upload the genome sequence of S.stipitis FASTA format in KAAS, and select the database of various strains with close relationship to it for two-way BLAST. ②IdentiCS is a fast independent genome annotation software, which adopts a simplified two-step annotation method and uses public databases such as Uniprot gene annotation information to identify more possible gene coding sequences than the gene sequences to be annotated. ③PSI-BLAST is a kind of iterative BLAST search. After the target microbial sequence is submitted online, the comparison result returned with the selected online database is characterized by increasing the number of similar sequences that can discover the evolutionary relationship of distant...
Embodiment 2
[0024] Features of the genome-scale metabolic network model of Pichia stipitis:
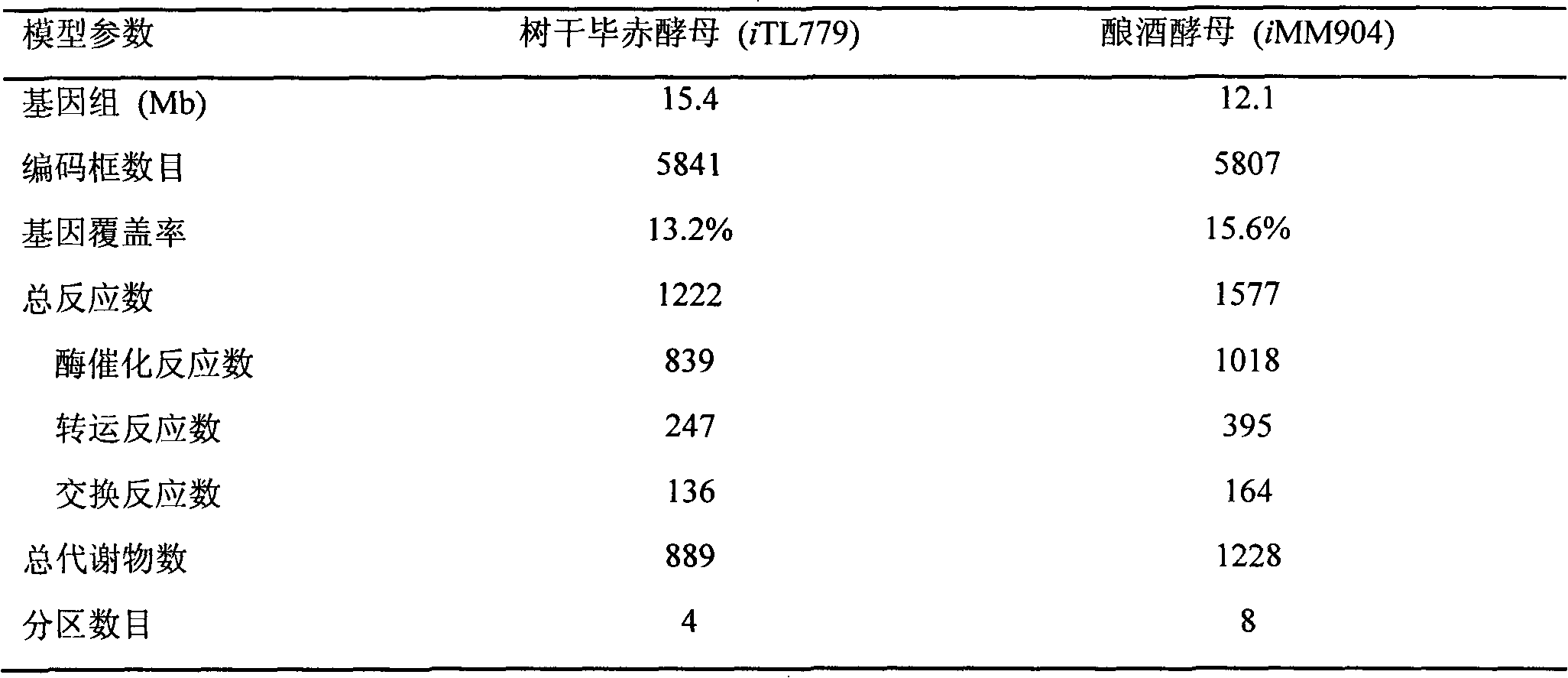
[0025] Using the above model construction method, we obtained a genome-scale metabolic network model of Pichia stipitis, including 1222 reactions, 889 metabolites and 779 genes, distributed in 4 subcellular systems, named iTL779. In order to confirm the built model, the typical parameters of iTL779 and the published Saccharomyces cerevisiae model iMM904 were calculated, and compared, it was found that the annotated genes with metabolic functions of the constructed Pichia stipitis iTL779 accounted for 13.2% of all Pichia stipitis genes. This data reasonably reflects that the gene annotation rate of eukaryotic cells is generally between 10% and 15%, and other parameters of the model are also within the normal range.
[0026] Table 2 Comparison of model features
[0027]
Embodiment 3
[0029] Validation and analysis of a genome-scale metabolic network model in Pichia stipitis:
[0030] Based on the flux balance analysis method based on Matlab, certain substrate absorption and other non-restrictive simulation conditions (such as nitrogen source, sulfur source, dissolved oxygen, etc.) are set to investigate different carbon sources (in cellulose hydrolyzate) with cell growth as the target equation. The five kinds of monosaccharides with the most content: D-glucose, D-mannose, D-galactose, D-xylose and L-arabinose) under the cell growth results, found that the maximum specific growth rate obtained by simulation and the actual measured by experiment The results are very close, thus confirming the accuracy of the constructed Pichia stipitis iTL779.
[0031] COBRA-Based Single-Gene, Single-Reaction Knockout Program Identifies Minimal Genes, Reaction Sets Respectively 110 Core Genes and 158 Cores Required to Sustain Pichia stipitis Cell Growth on Glucose as Carbon ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com