Method for eliminating PCR amplification product pollution based on IIs type restriction endonuclease
A technology for restriction endonuclease and amplification product, which is applied in the field of eliminating PCR amplification product contamination based on type IIs restriction endonuclease, and can solve problems such as affecting PCR amplification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
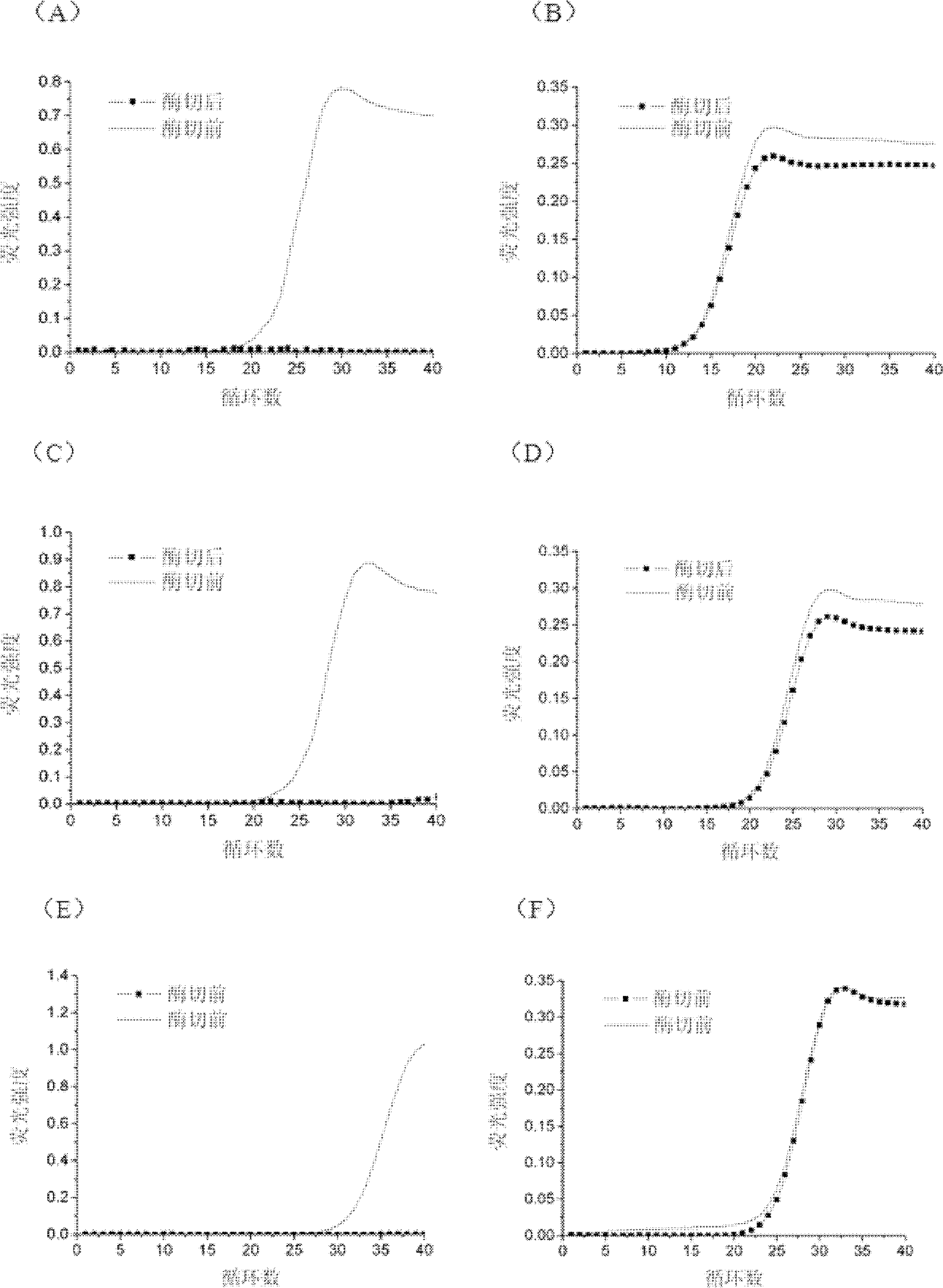
[0028] In order to investigate the elimination effect of this method on the product contamination after single-stranded template amplification, the single-stranded probe ligation product S-SRY was used as the single-stranded template, and the 10 μl ligation reaction system consisted of: genomic DNA extracted from human whole blood (100 ~ 200 ng), Probe 1 and Probe 2 mixture (5 fmol each), 1 U Ampligase, 20 mM Tris-HCl (pH 8.3), 25 mM KCl, 10 mM MgCl2, 0.5 mM NAD, 0.01% Triton X-100. The probe ligation conditions were pre-amplification at 94°C for 5 min, followed by 1 min at 94°C, and 4 min at 60°C, for a total of 10 cycles. The obtained ligation products were prepared by serial dilution after purification.
[0029] Design a pair of primers with the sequence of the single-stranded template S-SRY as the target sequence. The upstream and downstream primers match the sequence of the target gene, but the upstream primers are recognized by the FokI enzyme consisting of the recogniti...
Embodiment 2
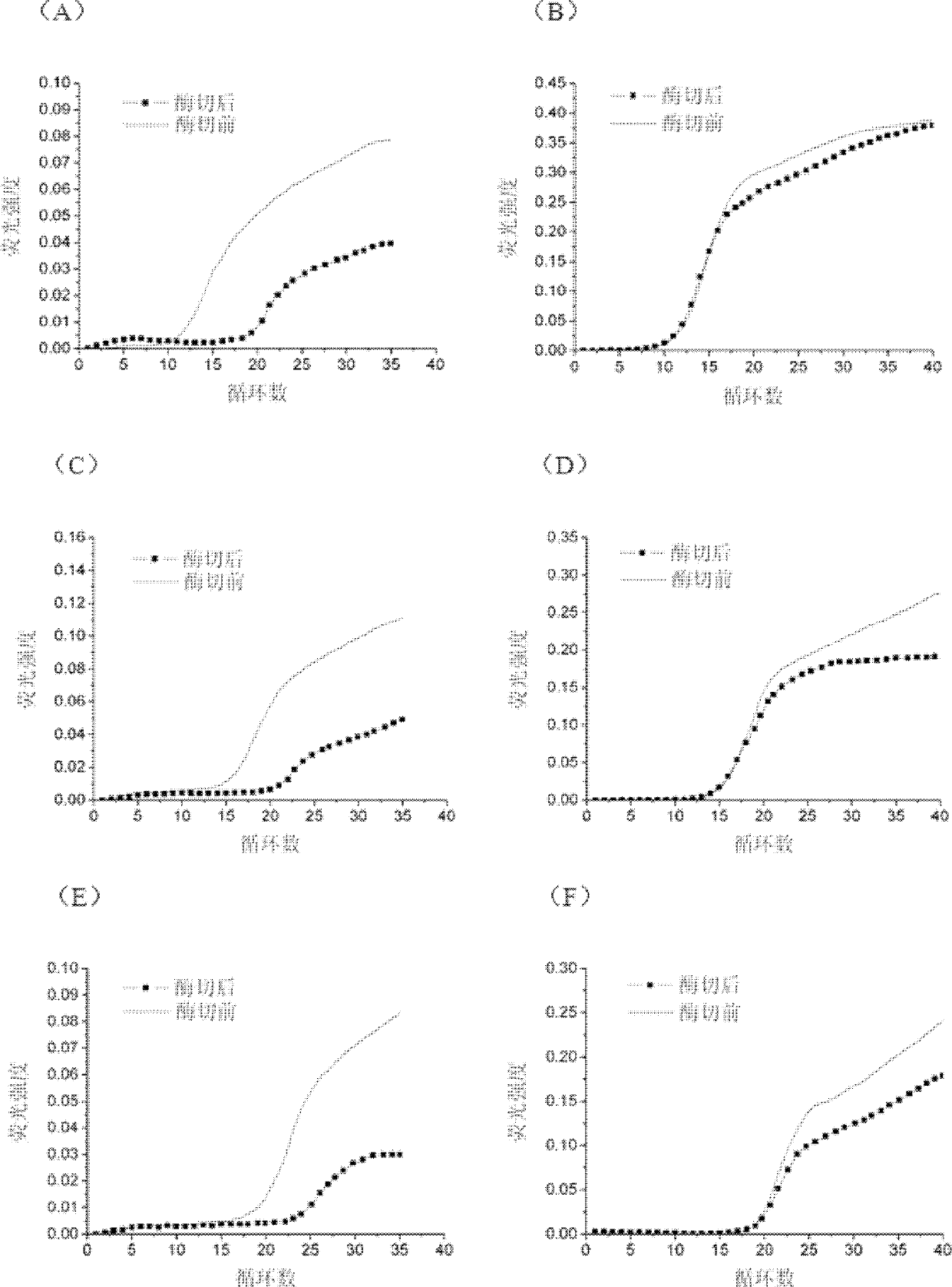
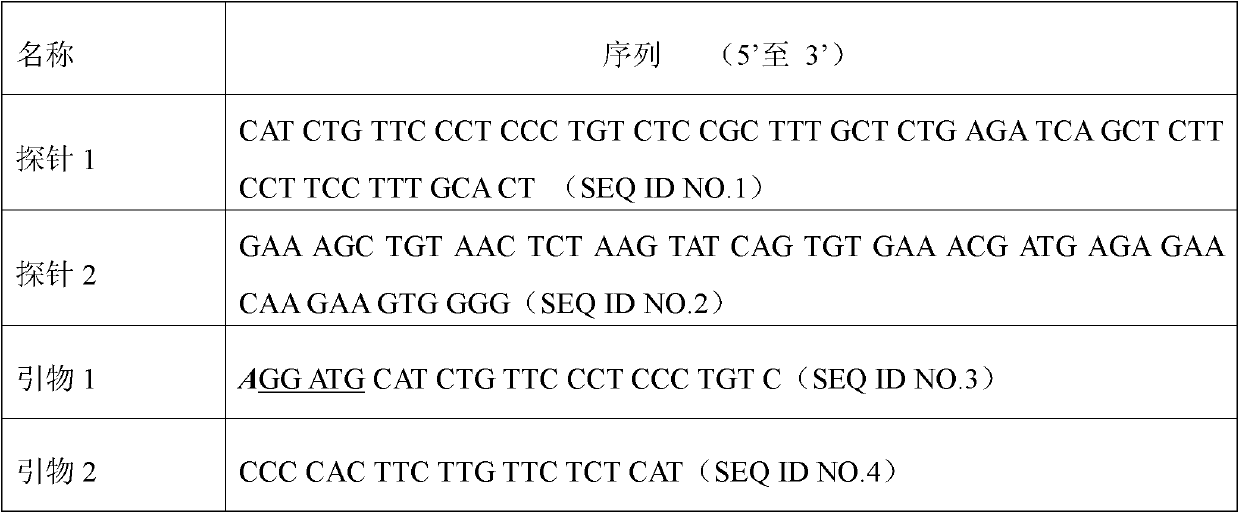
[0039] In order to investigate the elimination effect of the present invention on the product contamination after double-stranded template amplification, the short nucleic acid fragment PPAR-N is used as the target double-stranded template, and a pair of primers are designed. The upstream and downstream primers match the sequence of the target gene, but the upstream primer introduces Mismatched bases recognized by type IIs restriction enzymes. The templates and primer sequences of Example 2 are shown in Table 3, where the underlined bases are mismatched bases.
[0040] Table 3. Example 2 primer sequence
[0041]
[0042] The total volume of the PCR reaction system is 25 μL, and the PCR blank reaction system without DNA template is 2 mmol / L MgCl 2 , 0.5mmol / L dNTP, 1×PCR Buffer (100mmol / L Tris-HCl, 500mmol / L KCl, 1% X-100), 1U Taq enzyme, 10 pmol primer, 0.4×SYBG dye, 0.6U FokI enzyme.
[0043] The double-stranded template PPAR-N enzyme digestion condition is 37°C for 40...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com