Biomolecule affinity constant determination method based on DNA (Deoxyribonucleic Acid) origami
A measurement method and biomolecular technology, applied in the field of detection, can solve problems such as large errors, cumbersome steps, and complexity, and achieve the effects of high specificity, good repeatability, and less sample consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
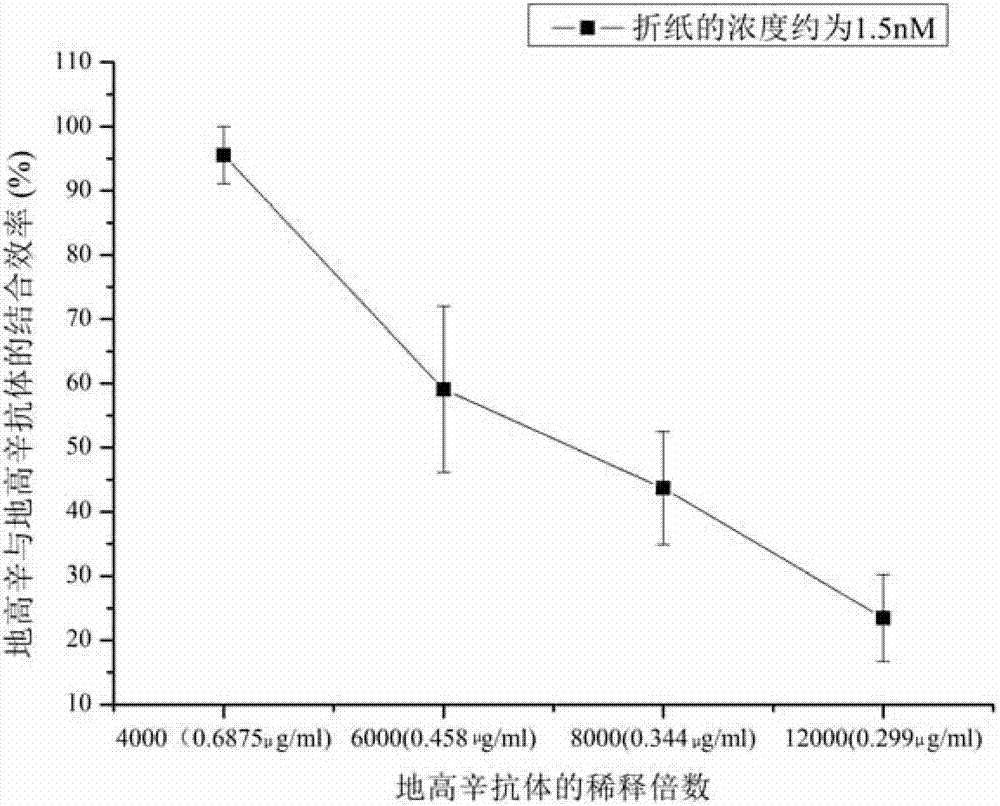
[0038] Example 1 Determination of digoxin and digoxin antibody affinity constant
[0039] 1. Preparation of digoxin-modified rectangular DNA origami
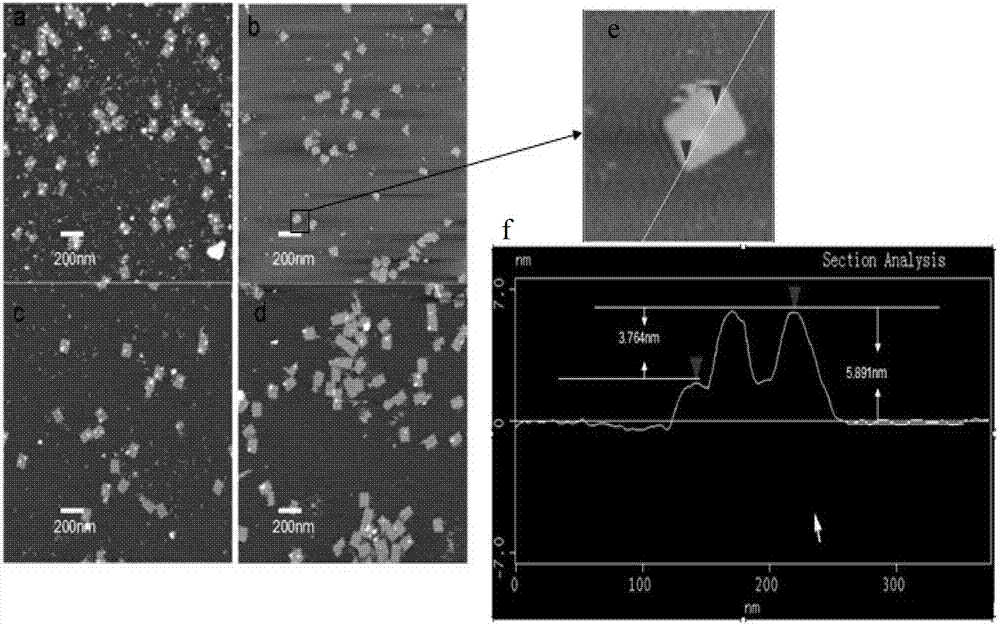
[0040] The DNA origami designed in this example utilizes M13mp18DNA and staple single-strand self-assembly to form a rectangular pattern with a length and width of 100 nm and 70 nm, respectively. Such as figure 1 Shown, at two positions on the DNA origami, the ends of the staple strands are modified with digoxigenin for binding to the digoxigenin antibody.
[0041] Mix M13mp18 DNA single strands and staple single strands (including digoxin-modified staple single strands) at a molar concentration of 1:10, and place in 1×TAE / Mg 2+ Buffer system (Tris 40mM, acetic acid 20mM, EDTA 2mM, MgCl21 2 .5mM, pH value 8), the total volume is 61μL, and then placed on the PCR instrument to anneal from 95°C to 20°C at an annealing speed of 0.1°C / 10s. After the reaction is completed, excess staples are removed by ultrafiltration Short chains...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com