Cutinase gene capable of efficiently producing cutinase and application thereof
A kind of cutinase and gene technology, applied in the field of cutinase gene, can solve problems such as high price, hindering the production of cutinase by recombinant bacteria, unsuitable for fermentation production and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
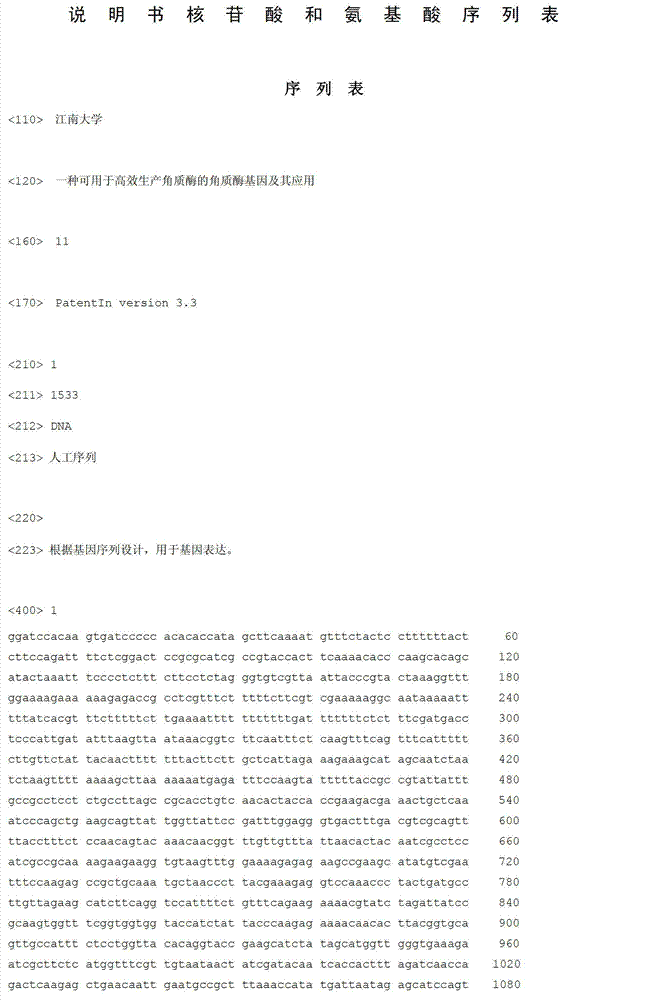
[0022] Embodiment 1 novel cutinase coding gene
[0023] A cutinase gene that can be used for high-efficiency production of cutinase, the nucleotide sequence of which is shown in SEQ ID NO.1. The method of obtaining it is to synthesize and optimize the gene encoding cutinase to obtain tfu, artificially add α-factor signal peptide and kozak sequence at the N-terminal, and clone it into pYES2.
Embodiment 2
[0024] Construction and identification of embodiment 2 recombinant yeast
[0025] Using Saccharomyces cerevisiae BY4741 as a gene template, four strong constitutive promoters: ADH1, HXT1, TEF1, and TDH3 were obtained according to the primers P1-P8 in Table 1, which were connected to the plasmid pYES2, and the inducible promoter GAL1 was used as a control to construct Four recombinant expression vectors were selected: pY-ADH1-tfu, pY-HXT1-tfu, pY-TEF1-tfu, pY-TDH3-tfu. The above four recombinant plasmids were transformed into S. cerevisiae INVsc1 competent cells by lithium acetate chemical method, and the colonies that could grow on the uracil-deficient YNB plate were successively transferred for three generations on the plate to obtain genetically stable recombinants. A number of positive recombinants were picked and verified by PCR with primers tfu-F and tfu-R, and a fragment of about 1095bp was obtained, indicating that the four recombinant plasmids had been successfully tra...
Embodiment 3
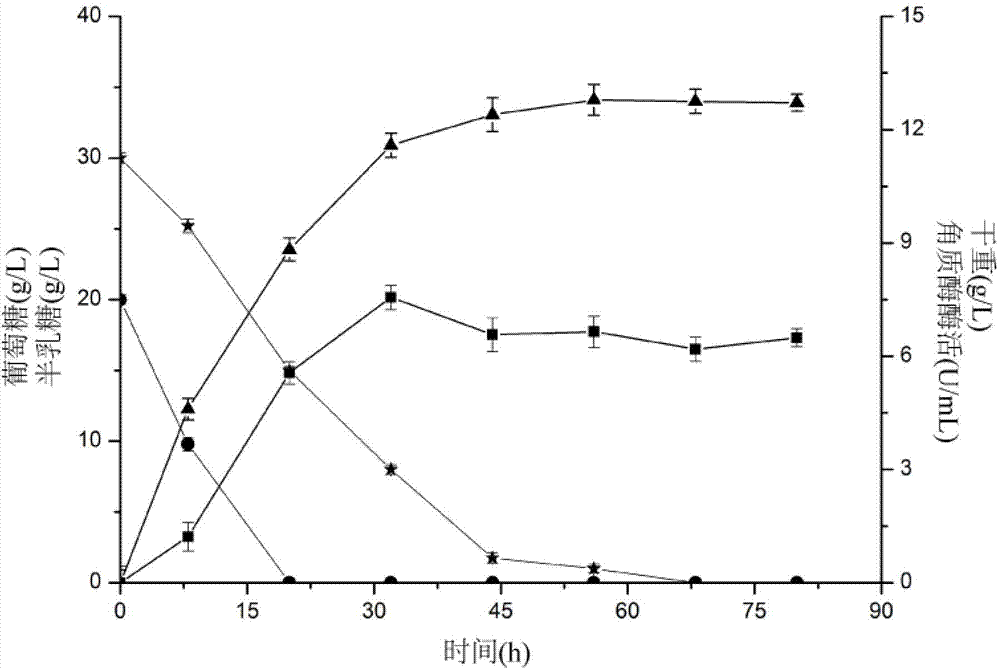
[0029] Example 3 Analysis and Comparison of Constitutive Promoter Reconstructed Saccharomyces Produced Cutinase
[0030] Inoculate the reconstituted strain into the seed medium, fill 50mL of a 500mL shaker flask with a temperature of 30°C, a rotational speed of 200r / min, and a culture time of 24-31h. According to the inoculum amount of 10%, the cultured seeds were inserted into the fermentation medium (initial sugar concentration was 20g / L), and cultivated under the conditions of 28°C and 200rpm. Compared with the control bacteria: TEF1p, TDH3p dry weight and enzyme All activities were improved; after 90 hours of fermentation, TEF1p had better dry weight and enzyme activity expression, which were 11.88U / mL and 17.85g·DCW / L, respectively. ADH1 had no effect, and the enzyme activity of HXT1 decreased instead.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com