Allglo probe-based detection method of anopheles sinensis knockdown resistance gene mutation site
The technology of a mutation site and detection method is applied in the detection of Anopheles sinensis knockdown resistance (kdr) gene mutation sites, and the field of fluorescence quantitative PCR detection of Anopheles sinensis knockdown resistance (kdr) gene mutation sites , which can solve the problems such as the need to improve the detection sensitivity and specificity, the complex synthesis of TaqMan probes, and the reduction of the test sensitivity and specificity, so as to reduce the background influence, improve the detection sensitivity and specificity, and improve the Tm value.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The design of embodiment 1 primer and probe
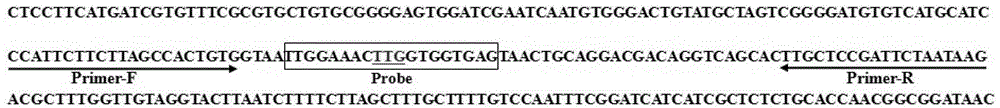
[0042] According to the Anopheles sinensis knockdown resistance (kdr) gene sequence (GenBank accession number GI84646709) and the L1014 site mutation, a pair of primers and three probes were designed with Primer Premier6.0: Primer 1 (upstream primer Primer-F ) and primer 2 (downstream primer Primer-R) nucleotide sequences are shown in SEQ ID NO: 1 and SEQ ID NO: 2, probe 1 (kdr-TTG), probe 2 (kdr-TTT) and probe 3 (kdr-TGT) nucleotide sequences are shown in SEQ ID NO: 3, SEQ ID NO: 4 and SEQ ID NO: 5. NEP gene markers, wherein the 7th base C and the 8th base T of the three probes are locked nucleotides. For the specific design regions of the above primers and probes, see figure 1 .
Embodiment 2
[0043] The design of embodiment 2 reaction system and condition
[0044] The invention adopts a triple probe detection method, that is, each reaction tube contains three probes kdr-TTG, kdr-TTT and kdr-TGT that can detect TTG, TTT and TGT.
[0045] Prepare the Allglo real-time fluorescent quantitative PCR reaction system: Roche Probe Master (2×) 5 μl, the final concentration of the upstream primer Primer-F and the downstream primer Primer-R is 500nM, the final concentration of the probes kdr-TTG, kdr-TTT and kdr-TGT is 400nM, Make up to 10 μl of sterilized water, one leg of Anopheles sinensis, and put it in LightCycler480 fluorescent quantitative eight-connected tube.
[0046] Determine the Allglo real-time fluorescent quantitative PCR reaction conditions: put the eight-tube tube in Roche Lightcycler480II for real-time fluorescent quantitative PCR amplification. The reaction conditions are set as follows: 95°C pre-denaturation for 5 minutes, 95°C denaturation for 15s, 60°C ann...
Embodiment 3
[0048] Embodiment 3 detection result judgment
[0049] Combined with the amplification curve to analyze and interpret the detection results.
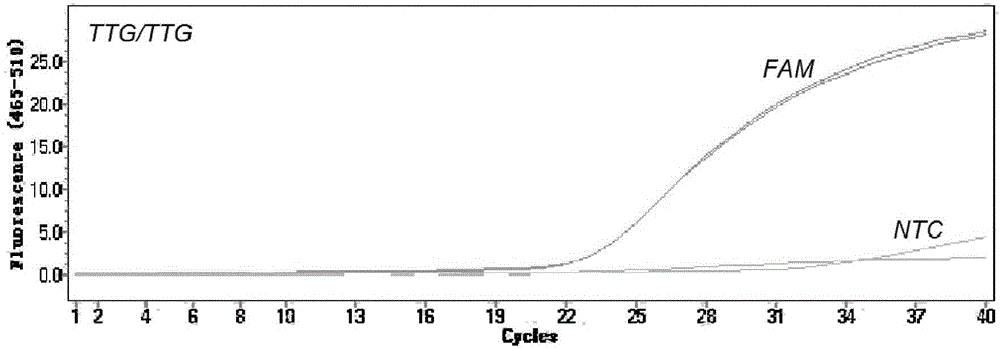
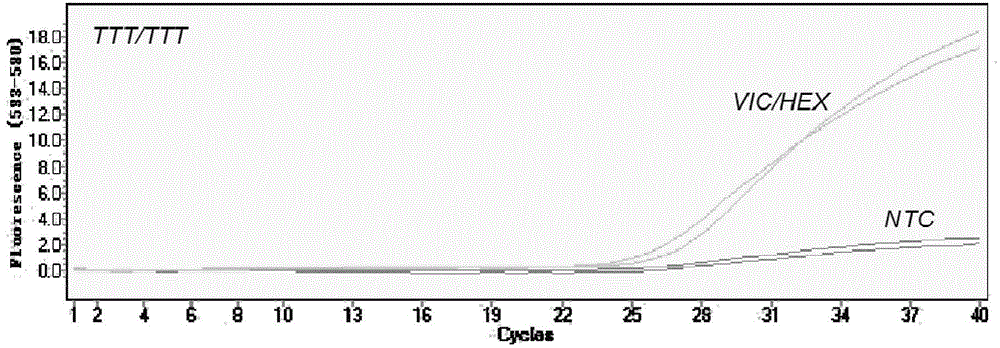
[0050] Kdr genotype identification: identify specific genotypes based on the fluorescence quantitative amplification curves of different channels. When only the FAM channel has an amplification signal, the kdr gene does not contain a mutation site, and it is a TTG homozygous wild type; when only the VIC / HEX channel has an amplification signal, the kdr gene has a L1014F mutation site, which is a TTT homozygous mutation type; when only the CY5 channel has an amplification signal, the kdr gene has the L1014C mutation site, which is a TGT homozygous mutant type; when any two of the FAM, VIC / HEX and CY5 channels have amplification signals, the kdr gene is Mutants (wild mutant heterozygous or mutation heterozygous), that is, when both FAM and VIC / HEX channels have amplification signals, but CY5 channels have no amplification signals, the kdr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com