Process for preparing an intermediate of sitagliptin via enzymatic conversion
A sequence and carrier technology, applied in the field of intermediates for the preparation of sitagliptin through enzymatic conversion, can solve problems such as consumption, ineffective and low-cost preparation, and adverse environmental effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0252] According to the present invention, the method for the preparation of formula (I) or any optically active (S) or (R) form thereof or an enantiomeric excess mixture of any of said forms can be carried out by various methods, including: using recombinant host cells, obtaining Desired enzymes from cell-free extracts / crude lysates of recombinant host cells, isolated from cell-free extracts / crude lysates or isolated from suitable organisms.
[0253] When the product is formed at the end of the reaction, it is then isolated from the reaction mixture using techniques well known in the art.
[0254] The (S) or (R)-3-hydroxyl-1-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a obtained above ]pyrazin-7(8H)-yl)-4-(2,4,5-trifluorophenyl)butan-1-one or its enantiomeric excess mixture is suitable as an intermediate for the preparation of sitagliptin body.
[0255] By reaction with methanesulfonyl chloride, (S)-3-hydroxy-1-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a...
Embodiment 1
[0263] Example 1 Cloning and gene expression analysis of chemically synthesized oxidoreductase and cofactor regenerating enzymes
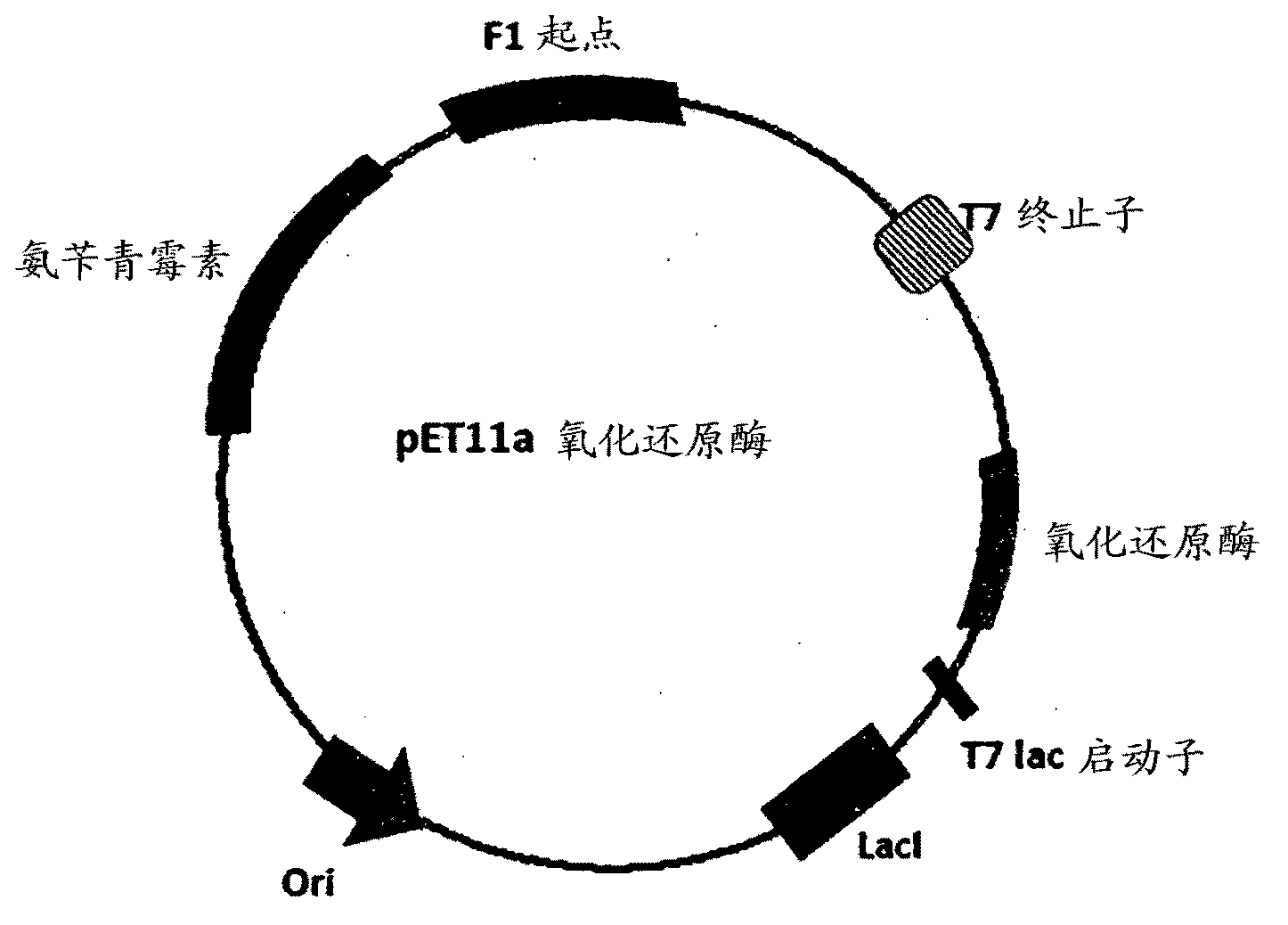
[0264] The DNA sequences deduced from the polypeptide sequences shown in Sequences Id No. 1, 2, 3, 4, 5 and 7 were codon optimized for expression in E. coli and cloned into the pET11a plasmid vector. In each case, the ligated DNA was further transformed into competent E. coli cells and the transformation mixture was plated on Luria agar plates containing ampicillin. Positive clones were identified based on their growth on petri plates as described above with ampicillin resistance and further restriction digests of plasmid DNA from them. Clones with the expected fragment lengths of the digested plasmid DNA samples were selected as putative positive clones. For each DNA sequence, one of such putative positive clones was subjected to nucleotide sequence analysis and found to have 100% homology to the sequence used for chemical synthesis. These pET1...
Embodiment 2
[0266] Example 2. Cloning and expression analysis of oxidoreductases derived from genomic DNA
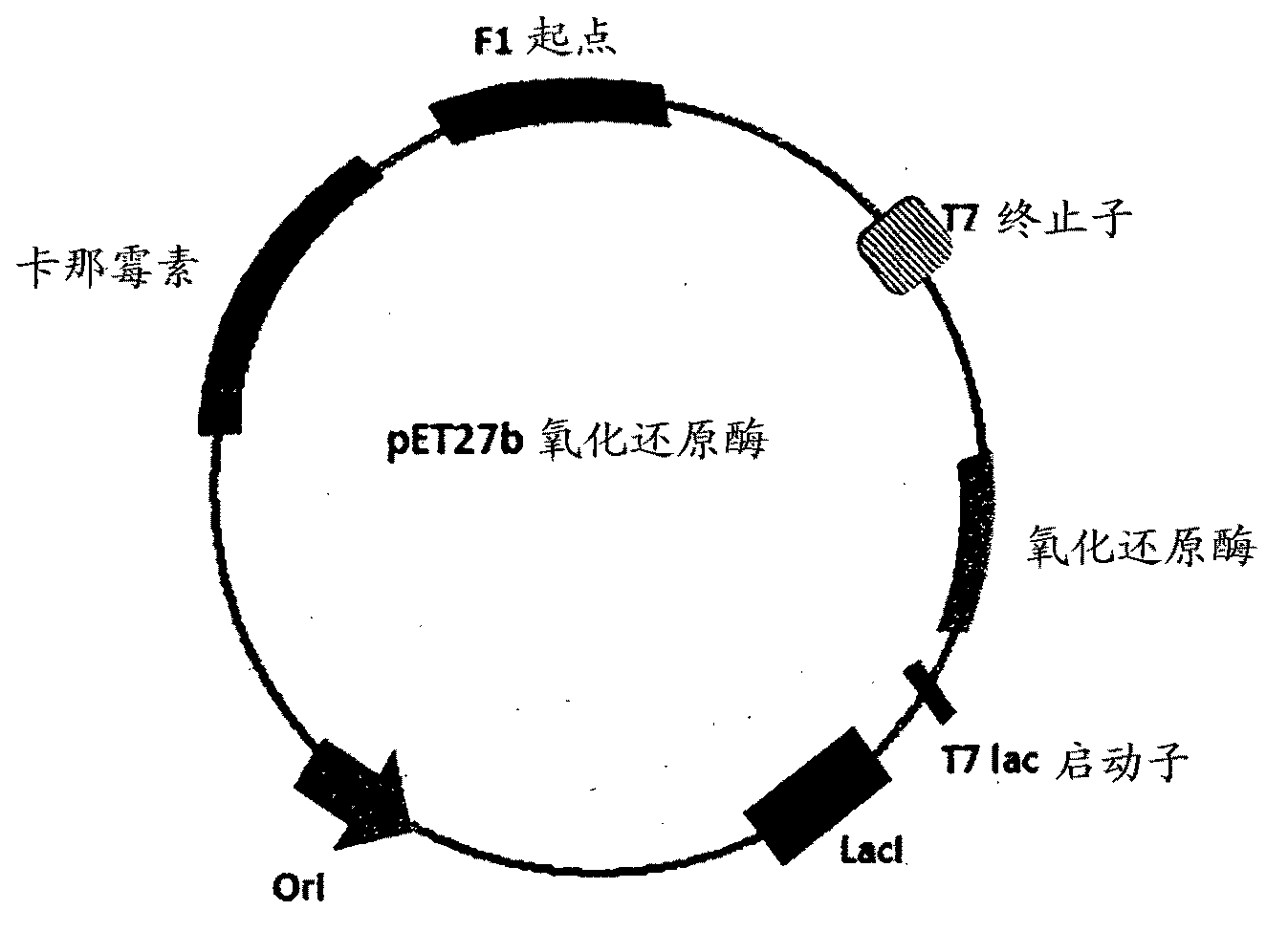
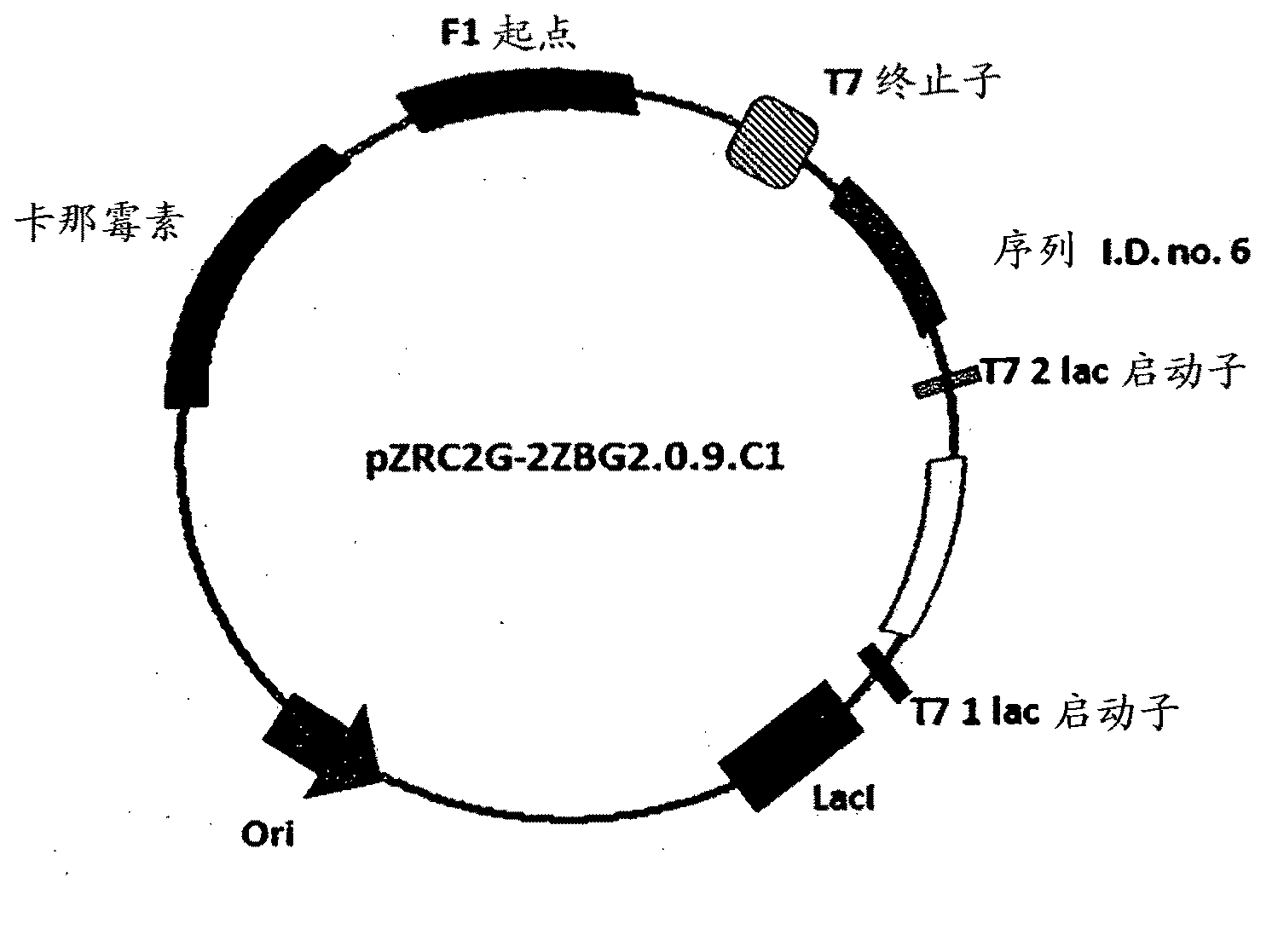
[0267] The DNA sequences deduced according to the polypeptide sequences shown in the sequence Id No.6, 8, 9, 10 and 13 of Table no.1 were amplified by PCR using the primers from Saccharomyces cerevisiae according to Table no.1B, and the DNA sequences from the large intestine The respective primers for Bacillus were PCR amplified with those of sequences Id No. 11 and 12 for expression in E. coli. These amplified PCR products were purified and subjected to restriction digestion with internal digestive enzymes to check the PCR products. PCR products of the correct band sizes corresponding to sequences Id No. 9, 11, 12 and 13 were subjected to restriction digestion with cloning enzyme NdeI-BamHI to be ligated to NdeI-BamHI digest vector pET27b, corresponding to sequences Id No. 6, PCR products of the correct band size for 8 and 10 were to be ligated into a blunt-ended vector digested...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com