Composition and method for site-specific recombination in hamster cells
A technology for hamster cells and host cells, applied in the field of genetic engineering, can solve problems rarely involved in screening stable and high-efficiency expression cell lines
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0336] Embodiment 1: Construction of pRu vector
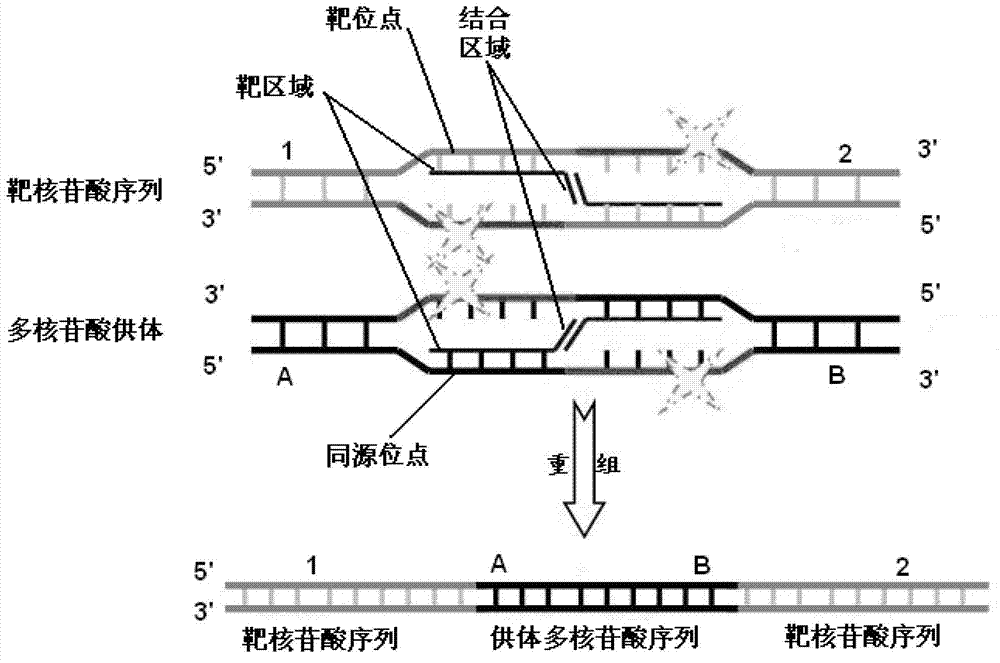
[0337] In this example, a pRu plasmid vector that can be used to insert homologous sites and foreign sequences (see Figure 11 , SEQ ID NO: 45). It contains three marker genes, RFP, ampicillin resistance gene and neomycin resistance gene, and promoters operably linked to these genes. When the homologous site is inserted, the pRu vector can be homologously recombined with the target site on the genome by the method of the present application, and the recombinant cells can be easily screened.
Embodiment 2
[0338] Embodiment 2: Construction of pCMV-RP vector
[0339] Using the genomic DNA of Escherichia coli as a template, the RecA gene was obtained by PCR using Phusion DNA polymerase (NEB Biolabs) and primers P1 and P2 as shown below:
[0340] P1: GACCGGCGCGCCGGATCCATGGCTATCGACGAAAACAAACAG (SEQ ID NO: 1);
[0341] P2: CACTGGACTAGTGGATCCTTAAAAATCTTCGTTAGTTTCTGCTACG (SEQ ID NO: 2).
[0342] The PCR product containing the RecA gene was cloned into the pROSE plasmid with the EGFP-puromycin-ubiquitin coding sequence removed ( Figure 12 , SEQ ID NO:46) at the BamH I restriction site, an expression vector containing the RecA gene was obtained and named pCMV-RP. The plasmid was sequenced to verify the insertion of the RecA gene. Amplify pCMV-RP to obtain the expression vector of the recombinase RecA, and purify it for future use.
Embodiment 3
[0343] Example 3: Construction of recombinant vectors containing iTS20 homology sites
[0344] In this example, the iTS20 high-expression site in the Chinese hamster ovary (CHO) genome was selected as the high-expression site, that is, the base sequence within 50 kb upstream or downstream of the Rpsa gene. use Figure 1t The Rpsa gene sequence in the mouse is searched for homology in the whole gene sequence of the mouse, and the mouse Rpsa gene homologous to the hamster Rpsa gene fragment is obtained, and then the sequence within 50kb upstream and downstream of the mouse Rpsa gene is known. According to the mouse DNA sequence within this range, primers are designed, and the genome material of the hamster is used as a template to amplify by PCR to obtain the sequence within 50kb upstream and downstream of the hamster Rpsa gene in the hamster genome. These sequences are the specific sequences of CHO-Genome.iTS 20, which can be used as high expression sites in the present inve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com