Lagerstroemia plant chromosomal in-situ hybridization method
An in situ hybridization and chromosome technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of difficult identification of chromosomes, difficult dyeing, blanks, etc., and achieve high hybridization specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] 1. Preparation of chromosome slide specimens:
[0050] At 9:00 in the morning, take the stem tip part of the innermost length of the shoot tip growth point of the annual branch of Lagerstroemia urophyllum with a length of 0.5 cm. Put them directly into the fixative solution (by volume, absolute ethanol: glacial acetic acid (analytical grade): chloroform (analytical grade) = 5:3:2) and fix in a -20°C refrigerator for 18 hours. Rinse and wash the fixative in distilled water, transfer to 0.075mol / L KCl solution and hypotonic for 30 minutes; rinse the growth point of the shoot tip after the pre-hypotonic treatment with distilled water, dry it with absorbent paper, and put it into the mixed enzyme solution Enzyme hydrolysis in a water bath at 37°C for 4 hours, and the mixed enzyme solution was prepared by 2.5% cellulase: 2.5% pectinase: 0.01% proteinase K in a volume ratio of 2:1:3. The slides were prepared by the flame drying method, and the microscopic examination was carri...
Embodiment 2
[0066] The difference from Example 1 is that the material is Yakushima crape myrtle. The steps and signal observation are the same as in Example 1.
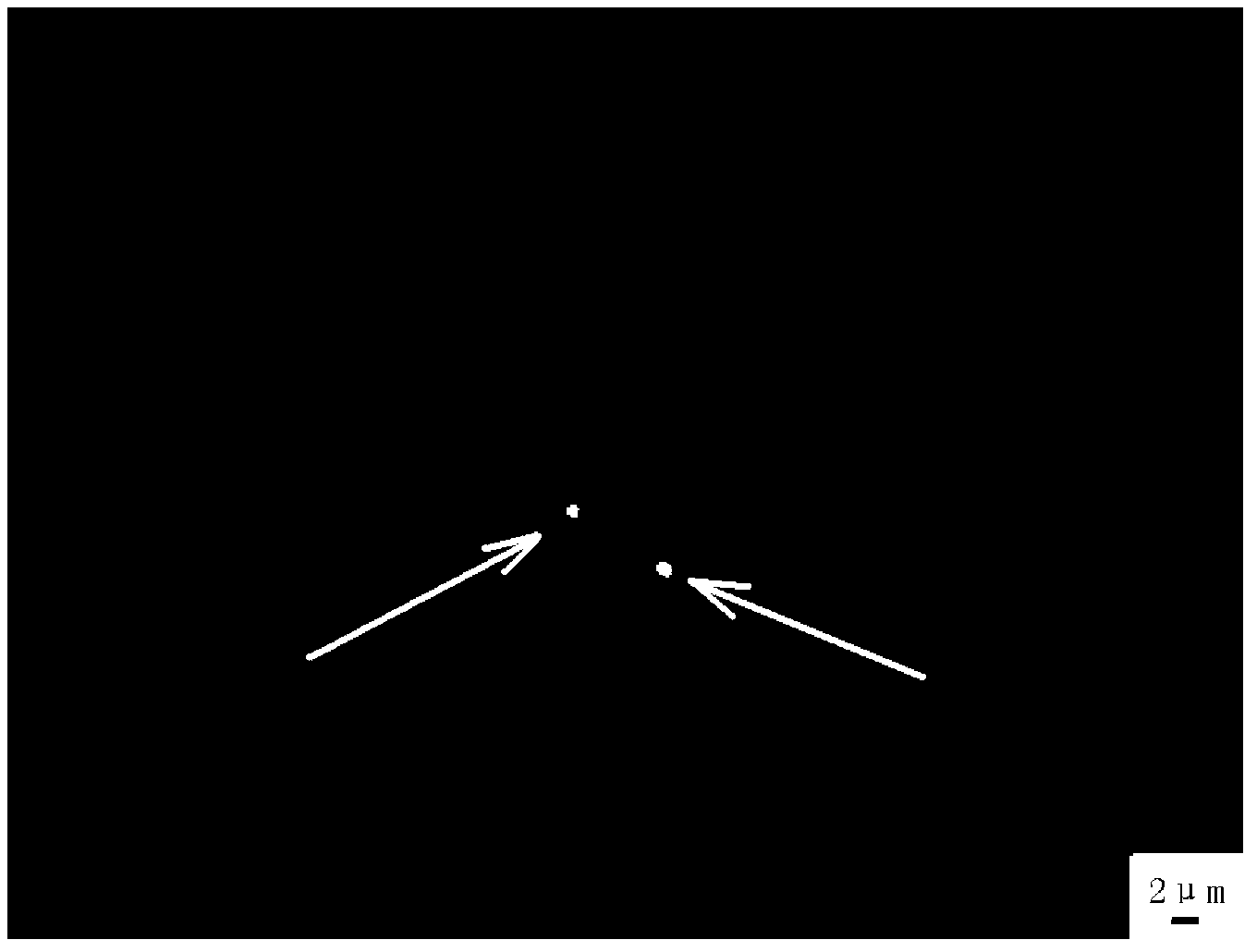
[0067] The results show that from figure 1 and figure 2 As can be seen in , there are 2 signal points (marked by arrows) in the hybridization of the Dig-labeled maize 45S rDNA probe to the Lagerstroemia genus chromosome preparation. The background interference is weak, and the sensitivity and accuracy are high, so that the in situ hybridization technique can be better applied to the chromosome detection of small chromosome plants such as Lagerstroemia indica, and provides a new way and method for the in-depth study of the chromosomes of Lagerstroemia genus plants.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com