Salmonella LAMP (loop-mediated isothermal amplification) primer group and kit and detection method
A Salmonella, primer set technology, applied in biochemical equipment and methods, microorganism-based methods, and microorganism determination/inspection, etc. High sensitivity, the effect of preventing the occurrence of false negative test results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] The establishment of embodiment 1 Salmonella LAMP detection kit
[0066] Salmonella LAMP detection kit, including LAMP primer set, LAMP reaction solution, Bst DNA polymerase, 10×SYBR, positive control, negative control and internal standard.
[0067] (1) LAMP primer set: Salmonella fimI was used as the detection gene and invA was used as the internal standard gene, and the online design software Primer Explorer version4 (http: / / primerexplorer.jp / e) was used to design LAMP primers.
[0068] The sequences of the fimI detection primers are as follows:
[0069] SafimI-F3: CACTAAATCCGCCGATCAA;
[0070] Safim I-B3: CAACGGTGAGGAGGTATTATC;
[0071] SafimI-FIP: TTCGGATCGCAGTCATTCAGGCGATTGGTGATACGACCG;
[0072] SafimI-BIP: GGTCAGGCAGATAACACCAACATTGCGGTGGTGCTATTATC;
[0073] Safim I-LF: TGGTGAAAGGCACCTGTG;
[0074] Safim I-LB: ATTTGCTGGCTGTCTCCTC.
[0075] The sequence of the invA internal standard primer set is as follows:
[0076] SainvA-F3: CGAAGCGTACTGGAAAGG;
[0077] ...
Embodiment 2
[0098] Embodiment 2 Sensitivity experiment
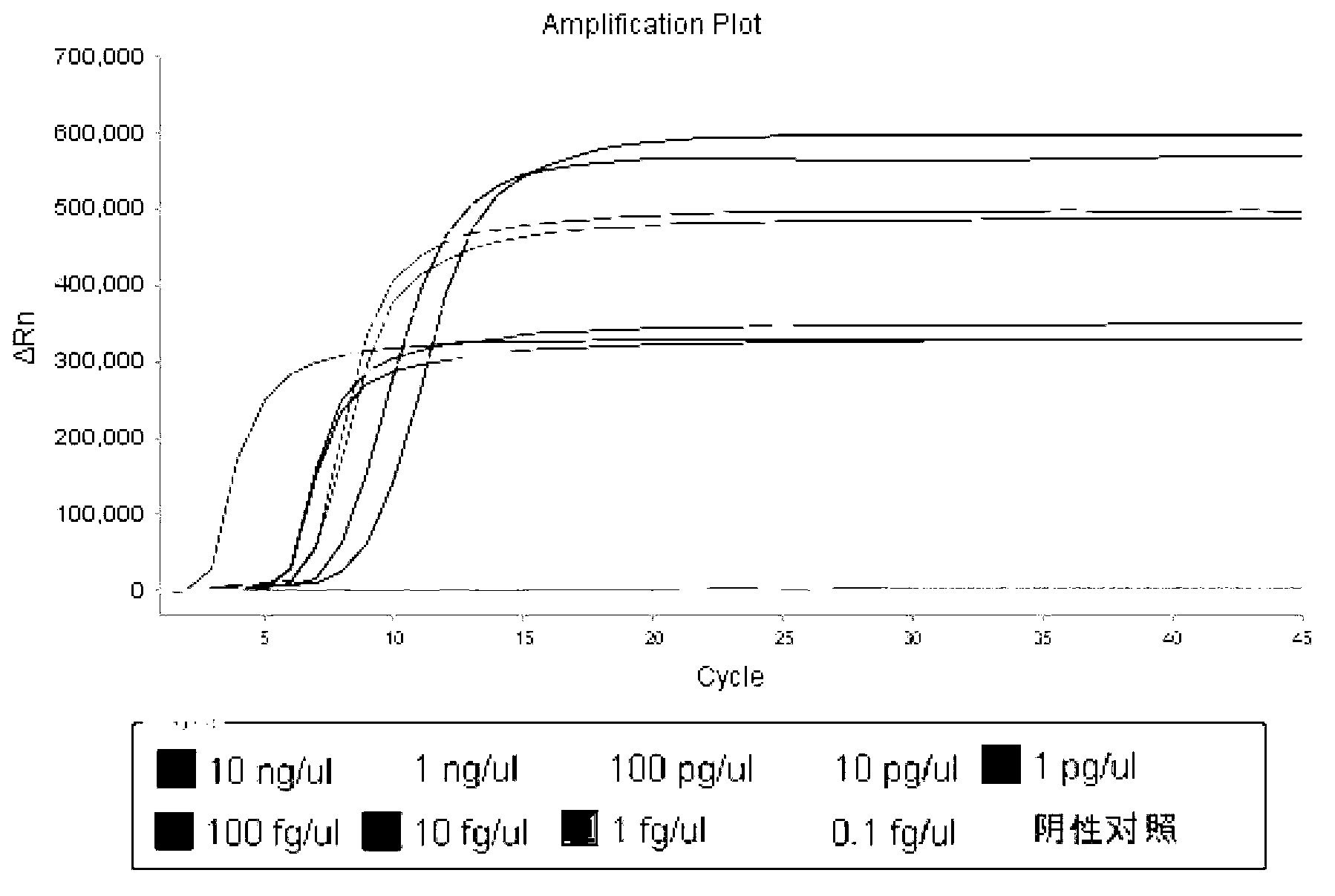
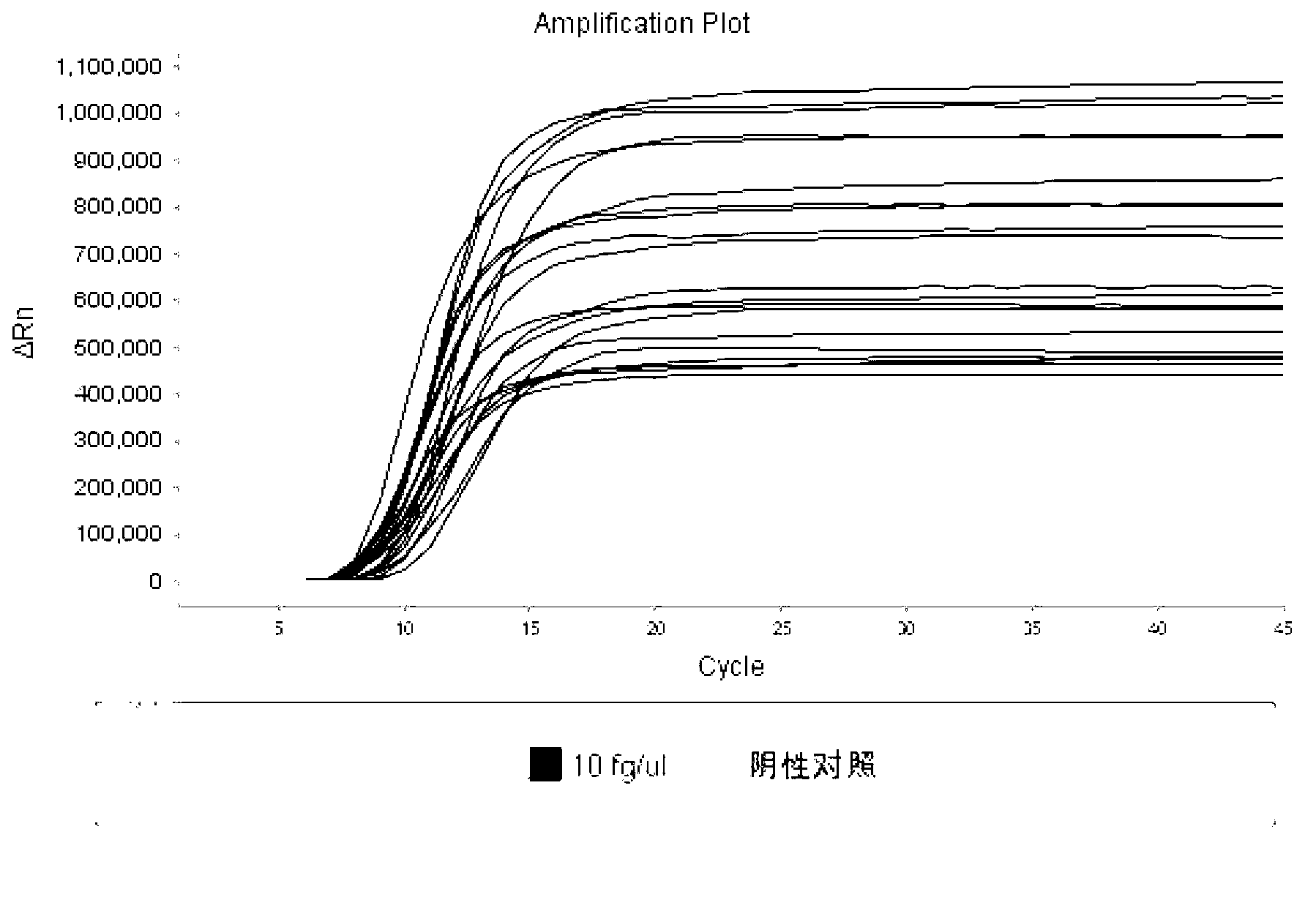
[0099] Use the internal standard constructed in Example 1 to conduct a sensitivity experiment. The plasmid was diluted 10 times as a quality control standard. The operation in Example 1 was used for detection, and the reaction was carried out at 63-65°C for 45 minutes. The minimum detection level of the internal standard gene was determined through the sensitivity experiment. Out limit 10fg / μl (such as figure 1 ), position the internal standard concentration at the concentration of the lowest detection limit ( figure 2 ).
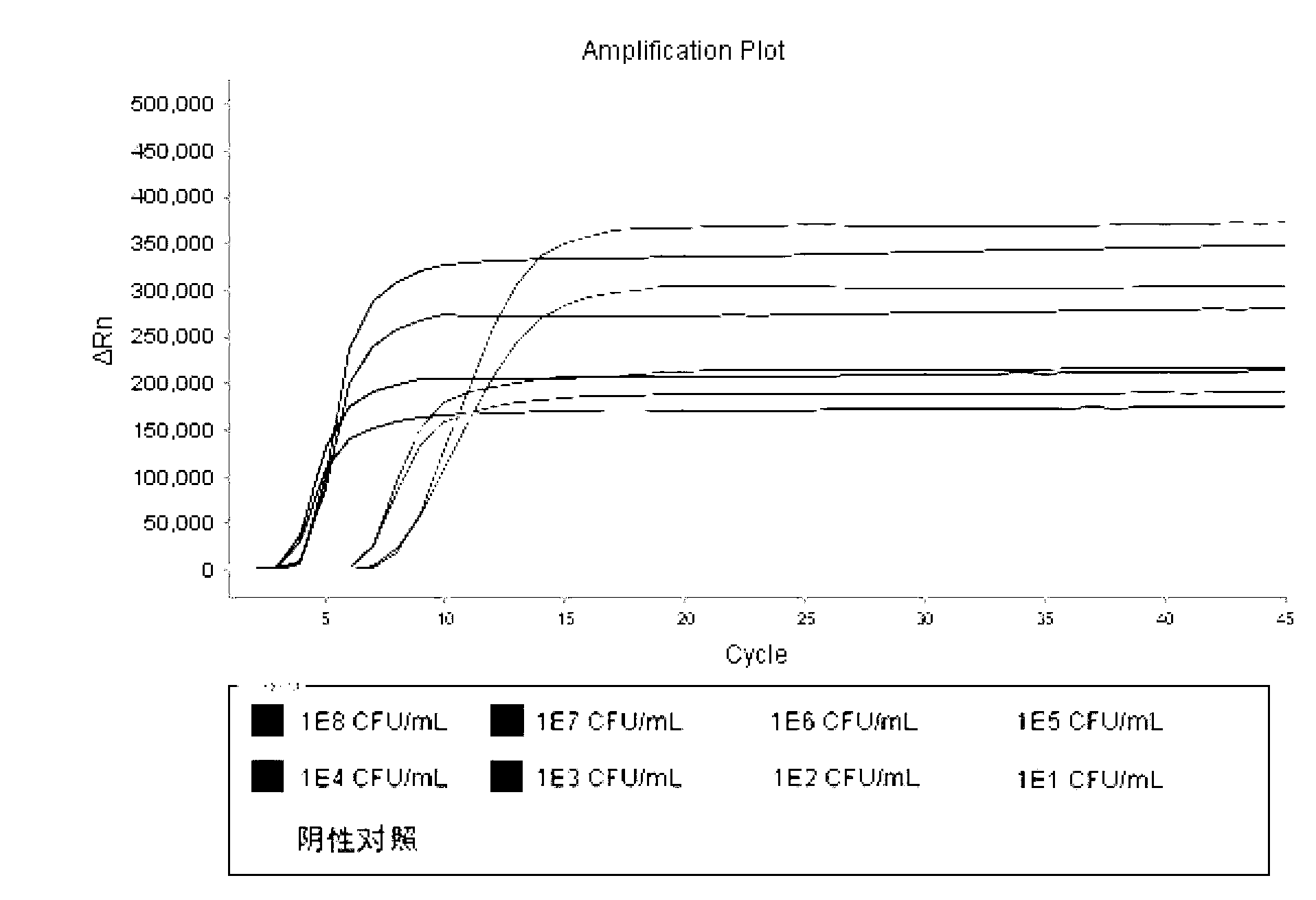
[0100] Cultivate the cultured Salmonella (refer to GB4789.4-2010), perform a 10-fold gradient dilution of the cultured bacteria, extract DNA according to the method in Example 1, and perform plate culture counting on the diluted bacteria solution, and plate The culture counting results were compared with the lowest sensitivity of this kit, the lowest detection degree of this kit was 2.4×10 3 CFU / mL, test resu...
Embodiment 3
[0101] Embodiment 3 specificity experiment
[0102] Using the method of Example 1 to culture Listeria monocytogenes, Staphylococcus aureus, Escherichia coli O157, Enterobacter sakazakii, Pseudomonas aeruginosa, Escherichia coli, Vibrio parahaemolyticus, Vibrio vulnificus, Abalone respectively Shigella sonnei, Shigella sonnei, Proteus vulgaris, Klebsiella, Micrococcus luteus extract, detect according to embodiment 1, 63 reaction 45min, the result is as follows Figure 4 It shows that the internal standard gene amplification of the positive control is normal, and it is effective for Listeria monocytogenes, Staphylococcus aureus, Escherichia coli O157, Enterobacter sakazakii, Pseudomonas aeruginosa, Escherichia coli, Vibrio parahaemolyticus, Vibrio vulnificus , Shigella baumannii, Shigella sonnei, Proteus vulgaris, Klebsiella, and Micrococcus luteus had no non-specific amplification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com