A kind of primer, method and kit for detecting hbv telbivudine resistance mutation
A drug-resistant mutation, telbivudine technology, applied in the field of medical testing, can solve problems such as failure to detect HBV polymerase region gene mutation in time, low success rate of hepatitis B virus sample amplification, delay in optimal treatment period, etc. Eliminates the link of conditional exploration, improves the success rate of amplification, and has the effect of high degree of automation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] A primer for detecting HBV telbivudine drug-resistant mutation, the design of the primer is two pairs of specific amplification primers designed for regions with high homology among different subtypes of HBV, including:
[0053] (i) Specific outer amplification primers:
[0054] SEQ NO1: GACTCGTGGTGGACTTCTCTC;
[0055] SEQNO2: CKTTGACATACTTTCCAATCAA; wherein K represents G or T;
[0056] (ii) Specific inner amplification primers:
[0057] SEQ NO 3: TCGTGGTGGACTTCTCTCAATT;
[0058] SEQ NO 4: TTCGTTGACATACTTTTCCAATCA;
[0059] The specific inner amplification primers SEQNO3 and SEQNO4 can also be used as sequencing primers for Sanger sequencing.
[0060] The method for detecting the HBV telbivudine drug-resistant mutation using the above primers comprises the following steps:
[0061] (i) Extract the HBV DNA in the serum of the sample.
[0062] (ii) Using HBV DNA as a template, perform the first round of PCR amplification according to the specific outer amplificatio...
Embodiment 2
[0080] Embodiment 2: detection method
[0081] 1. Use the conventional phenol / chloroform extraction method to extract the HBVDNA in the sample serum.
[0082] 2. The first round of PCR amplification method is as follows:
[0083] The total PCR system for one sample is 10 μl: 10*PCRBuffer 1 μl, dNTP 1 μl, primers SEQNO 10.4 μl, SEQNO 20.6 μl, sterilized water 5.8 μl, Taq enzyme 0.2 μl, HBV DNA template 1 μl.
[0084] The first round of PCR reaction procedure:
[0085]
[0086] 3. The second round of PCR amplification method is as follows:
[0087] The total PCR system for one sample is 20 μl: 10*PCRBuffer 2 μl, dNTP 2 μl, primers SEQNO3 1.2 μl, SEQNO4 1 μl, sterilized water 12.3 μl, Taq enzyme 0.5 μl, first-round product 1 μl.
[0088] The second round of PCR reaction procedure:
[0089]
[0090] 4. Enzyme purification of PCR products. The enzyme purification system for a sample is 11 μl: calf alkaline phosphatase (CIAP) 0.1 μl, exonuclease Exo I 0.5 μl, sterilized...
Embodiment 3
[0097] Embodiment 3: sample detection
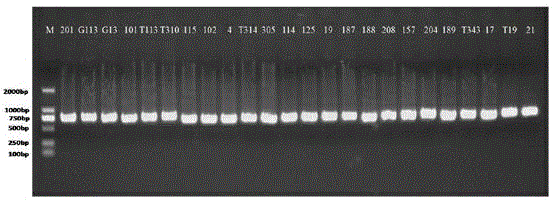
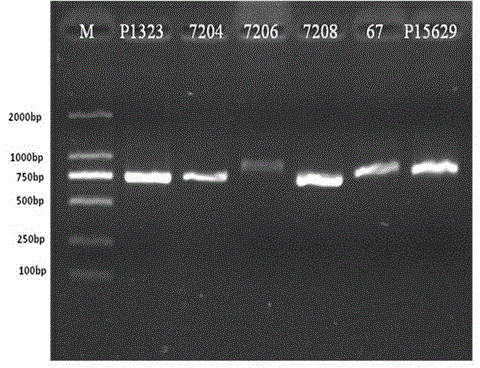
[0098] 30 samples of HBV infection were detected according to the methods of Examples 1 and 2. Results showed that all samples were successfully amplified (see Figure 2-3 ), and all samples were successfully sequenced to obtain the gene information of 180 and 204 sites (see Table 1).
[0099] For those skilled in the field of biomedicine, according to the sequencing map (attached Figures 6 to 11 ) to determine whether the new primers for detecting the mutation information at positions 180 and 204 are effective.
[0100] Table 1
[0101] sample number rtL180M rtM204I rtM204V 201 none none none G113 have have none G13 have none have 101 have none have T113 none have none T310 have none have 115 have none have 102 none have none 4 have have none T314 none none none 305 none have none 114 have none ha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com