Method for extracting plant deoxyribonucleic acid (DNA) and special kit thereof
A kit and plant technology, applied in the field of extracting plant DNA, can solve the problems of limited use and poor extraction effect, and achieve the effects of good versatility, low cost and high yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
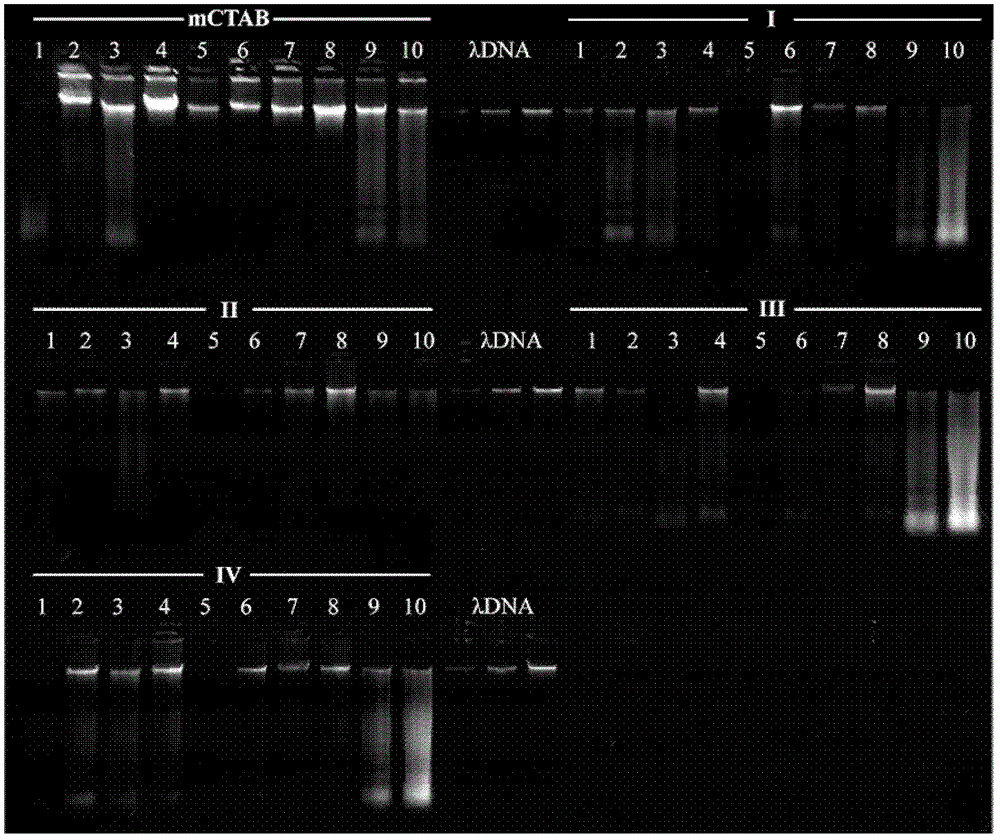
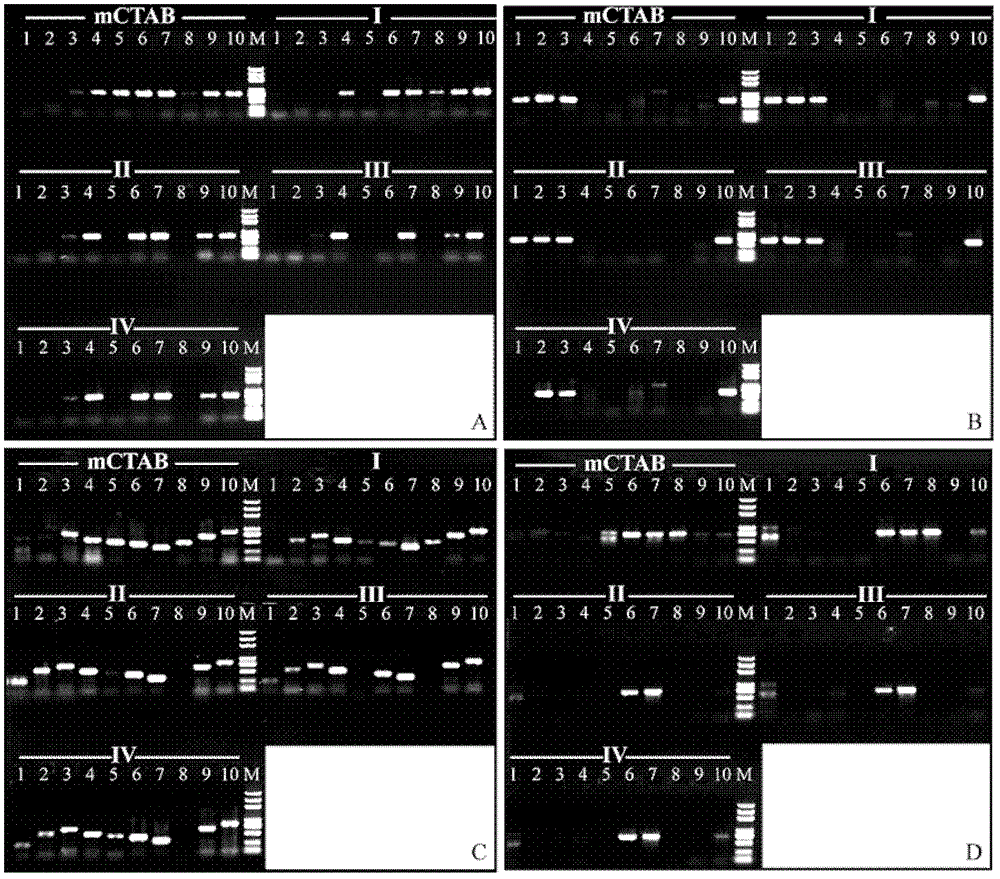
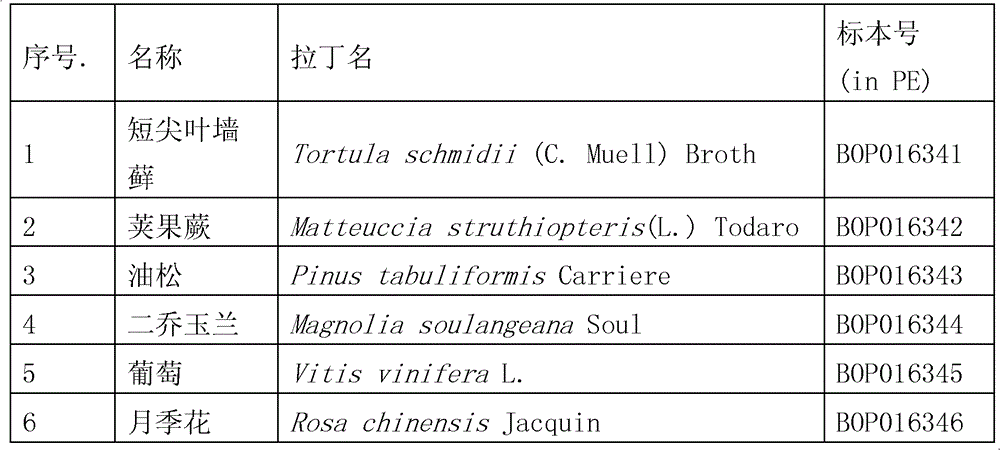
[0042] Embodiment 1, the method for extracting plant DNA of the present invention (hereinafter referred to as mCTAB method)
[0043] 1. Weigh 20mg of plant dry material, add quartz sand, grind it into powder, and transfer the powder to a 2.0mL centrifuge tube.
[0044] 2. Add 1mL of pre-cooled buffer A, mix well and then ice-bath for 15min; invert and mix 2-3 times during the ice-bath. Centrifuge at 7000×g for 10 min and discard the supernatant.
[0045] 3. Repeat step 2 until the supernatant is not viscous.
[0046] 4. Add 0.7mL buffer B (Table 3), mix well and then bathe in 65°C water bath for 90-120min, invert and mix several times during the water bath process. Centrifuge at 10000×g for 10min, and aspirate the supernatant into a new 2.0mL centrifuge tube.
[0047] 5. Add 0.7 mL of chloroform isoamyl alcohol solution (chloroform: isoamyl alcohol = 24:1), invert and mix for 10 min. Centrifuge at 10000×g for 10min, and aspirate the supernatant into a new 1.5mL centrifuge ...
Embodiment 2
[0059] The method is basically the same as in Example 1, the differences are as follows:
[0060] Buffer A is composed of solute and solvent; the solute and its final concentration in buffer A are as follows: disodium edetate 4mmol L -1 , NaCl 0.2M, polyvinylpyrrolidone 1g / 100ml; 1M Tris-HCl buffer solution with pH 8.0 8ml / 100ml; the solvent is water.
[0061] Buffer B is composed of solute and solvent: solute and its final concentration in buffer B are as follows: 1M Tris-HCl buffer solution with pH=8.0 8ml / 100ml, disodium edetate 20mmol L -1 , NaCl 1.2M, CTAB 2.5g / 100ml, sodium metabisulfite 0.5g / 100ml, sodium ascorbate 0.5g / 100ml, polyvinylpyrrolidone 1g / 100ml, β-mercaptoethanol 80ul / 100ml; the solvent is water.
[0062] The time of the ice bath was 10 minutes; the condition of the water bath was 90 minutes at 60°C.
Embodiment 3
[0064] The method is basically the same as in Example 1, the differences are as follows:
[0065] Buffer A is composed of solute and solvent; the solute and its final concentration in buffer A are as follows: EDTA disodium 6mmol L -1 , NaCl 0.3M, polyvinylpyrrolidone 3g / 100ml; 1M Tris-HCl buffer solution with pH 8.0 12ml / 100ml; the solvent is water.
[0066] Buffer B is composed of solute and solvent: solute and its final concentration in buffer B are as follows: 1M Tris-HCl buffer solution with pH=8.0 12ml / 100ml, disodium edetate 30mmol L -1 , NaCl 1.6M, CTAB 3.5g / 100ml, sodium metabisulfite 1.5g / 100ml, sodium ascorbate 1.5g / 100ml, polyvinylpyrrolidone 3g / 100ml, β-mercaptoethanol 120ul / 100ml; the solvent is water.
[0067] The time of the ice bath was 20 minutes; the condition of the water bath was 120 minutes at 70°C.
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com