PCR primer design method for identification of closely related species
A technology for primer design and species identification, applied in biochemical equipment and methods, microbial determination/inspection, calculation, etc., can solve the problems of poor primer specificity, poor sensitivity, and inability to effectively distinguish two species.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
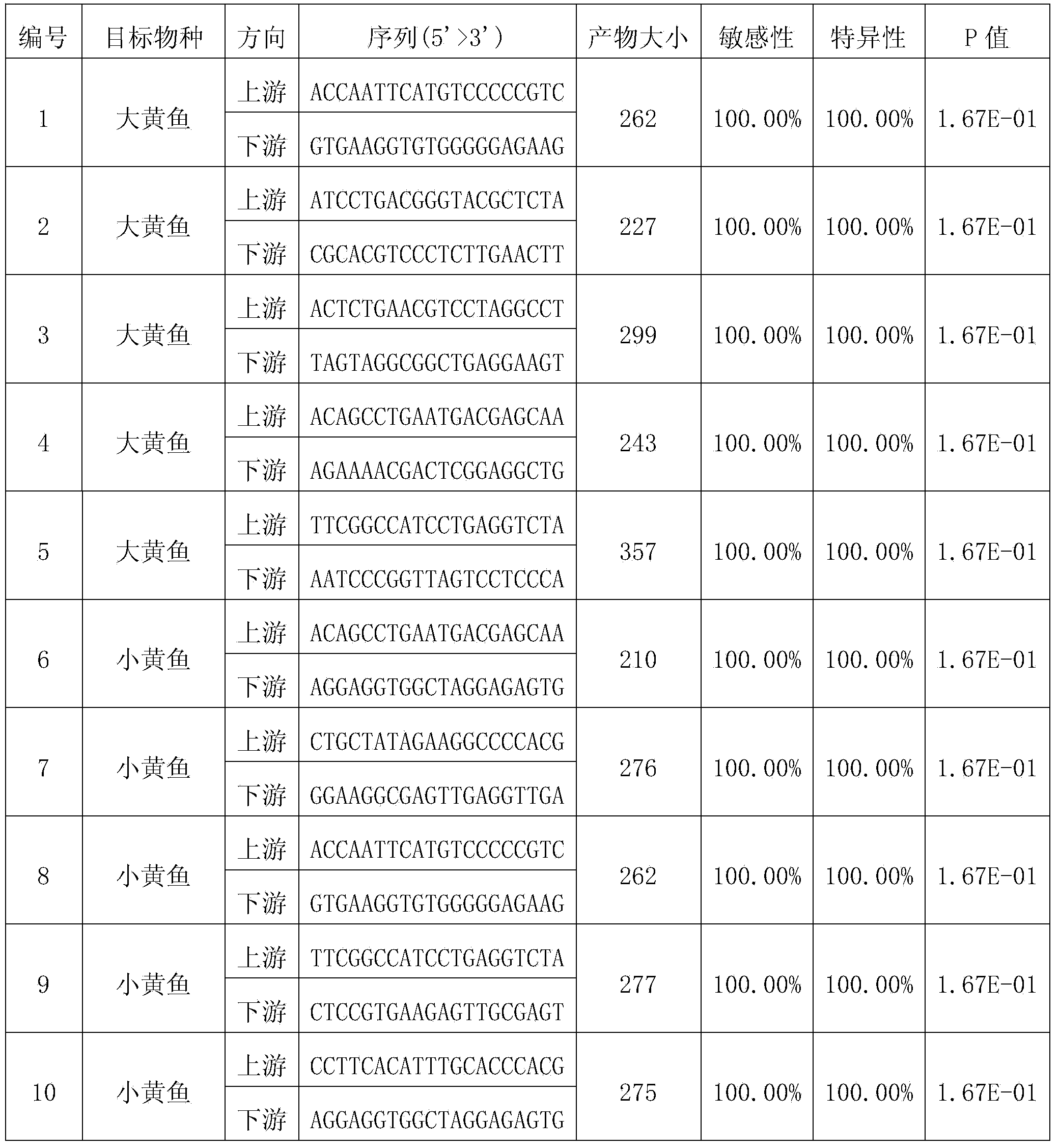
[0028] Example 1: A PCR primer design method for identification of closely related species large yellow croaker and small yellow croaker
[0029] Design PCR primers for identifying the target species of large yellow croaker and non-target species of small yellow croaker. Two pairs of specific primers can be designed, one pair specifically recognizes the mitochondrial DNA of large yellow croaker, and the other pair specifically recognizes the mitochondrial DNA of small yellow croaker; Design a pair of PCR primers that specifically recognize the mitochondrial DNA of large yellow croaker.
[0030] (1) Design of primers for specific recognition of large yellow croaker
[0031] 1. Obtain the sequence of the same gene in the target species and non-target species in the related species, and perform multiple sequence alignment to obtain the multiple sequence alignment file
[0032]Large yellow croaker and small yellow croaker are fishes with very close kinship. Mitochondrial DNA has ...
Embodiment 2
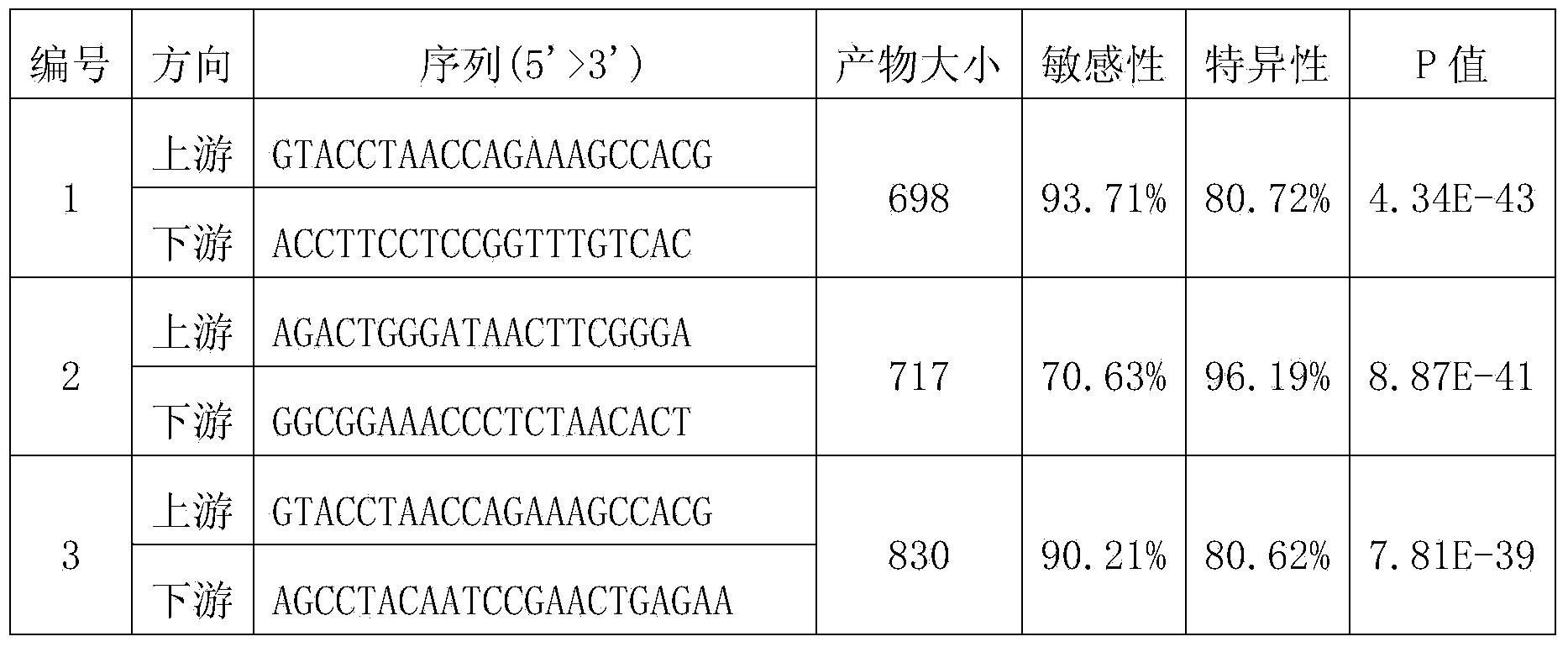
[0091] Example 2: Design of PCR primers for the identification of Bacillus bacteria in environmental samples
[0092] (1) Design of primers for specific recognition of Bacillus bacteria
[0093] 1. Obtain the sequence of the same gene in the target species and non-target species in the related species, and perform multiple sequence alignment to obtain the multiple sequence alignment file
[0094] The gene sequence of the target species Bacillus bacterium in the multiple sequence alignment file used is its 16S rDNA. The gene sequence of the non-target species is the 16S rDNA sequence of other bacteria of the Bacillus family except the bacteria of the genus Bacillus. These gene sequences are downloaded from the RDP database (http: / / rdp.cme.msu.edu / ), and there are some restrictions on the quality of the sequences when downloading, including: the sequence source strain must be a standard strain; the sequence source strain is an isolate; The sequence length must be greater than ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com