Osa-miR396d responding to rice black streaked dwarf virus infection, and application and production method thereof
A technology for dwarfing black stripes and viruses in rice, applied in the fields of applications, botanical equipment and methods, DNA/RNA fragments, etc., can solve problems such as no clear answer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
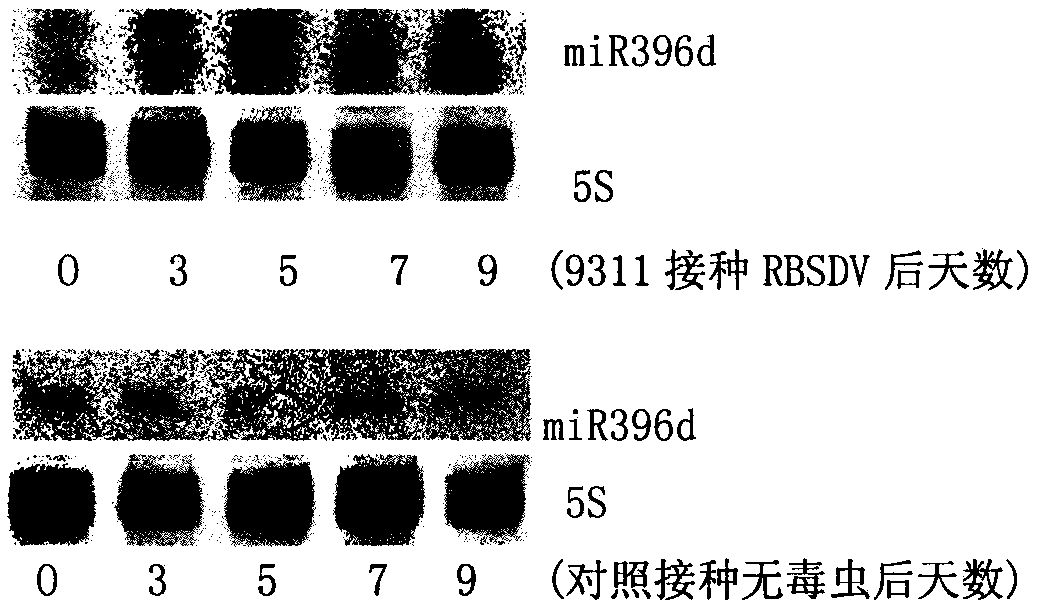
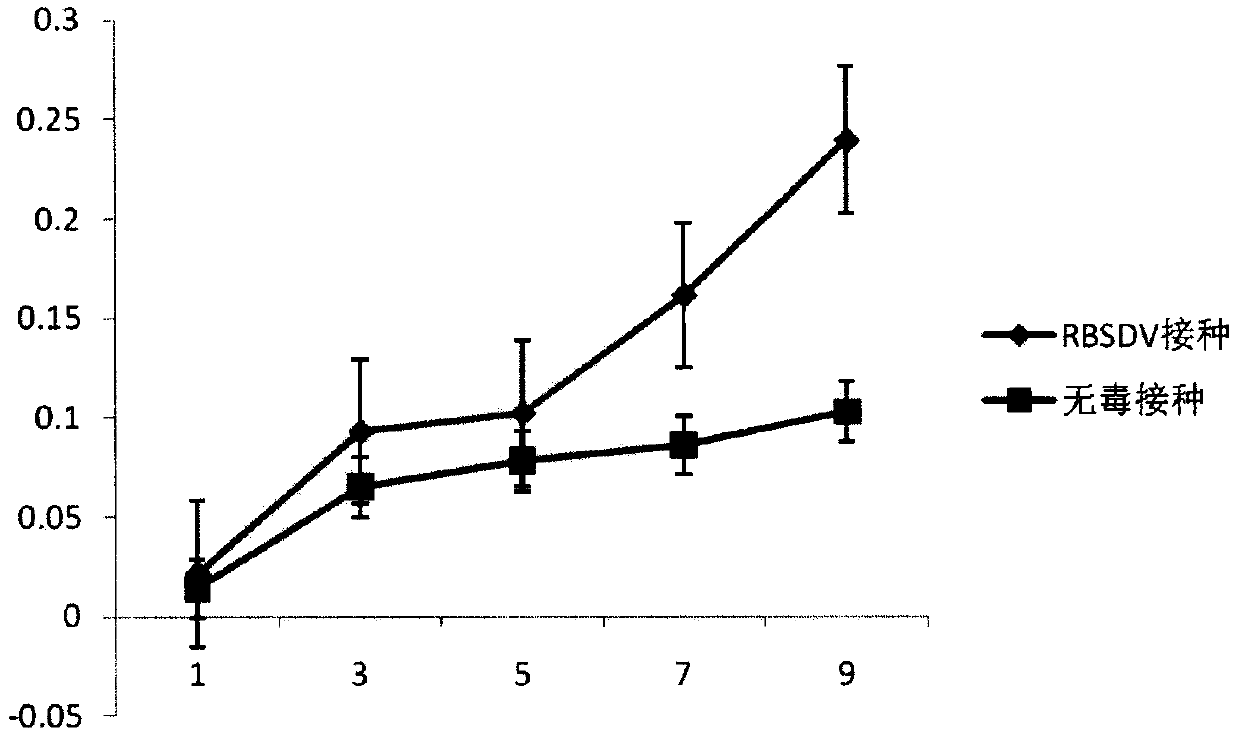
[0017] Embodiment 1, the discovery that osa-miR396d responds to virus infection
[0018] 1. Treatment of rice plants inoculated with rice black-streaked dwarf virus
[0019] The rice seeds were soaked at 25°C for 24 hours, and then germinated for 24 hours. After germination, the rice was planted in a 25°C incubator and the conditions were set as follows: temperature 28°C / 21°C (day 16h / night 8h), light intensity 400μmol / m2·s, relative humidity 70%, and nutrient soil to provide the growth conditions for rice. all the nutrients needed. When the rice seedlings grow to the 4-leaf stage, they are inoculated with SBPH with rice black-streaked dwarf virus, and the rice seedlings inoculated with non-toxic SBPH are used as the control; after 2 days, the SBPH are driven out, and the inoculated seedlings are Transplant to the greenhouse (25°C), and take samples when the diseased plants show dwarf symptoms after 3 weeks.
[0020] 2. Discovery of osa-miR396d
[0021] 1. RNA extraction ...
Embodiment 2
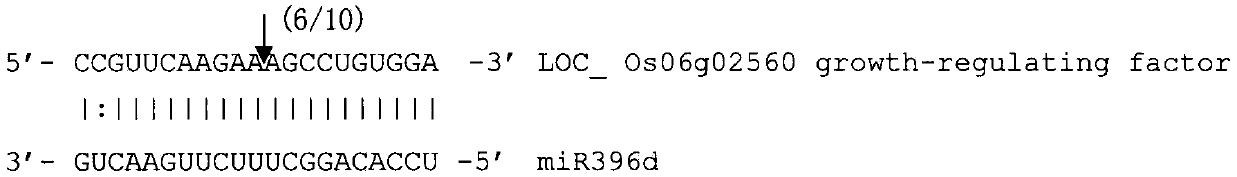
[0048] Example 2. Prediction and verification of target genes of miRNAs in response to rice black-streaked dwarf virus infection
[0049] Since plant miRNAs are nearly completely complementary to target gene mRNAs, the target genes of osa-miR396d can be predicted by bioinformatics methods. Use the psRNATarget program to search for a cDNA or gene that is nearly completely complementary to the miRNA sequence in the rice full-length cDNA library http: / / plantgrn.noble.org / psRNATarget / , which is the target of the miRNA; the parameters are set to: Patscan program parameters are default Set the maximum number of mismatches allowed to be 3, the 10th and 11th bases of miRNA are not allowed to have mismatches, and the known functional genes with the highest homology are used for annotation.
[0050] See Table 1 for the prediction results.
[0051] Table 1. Target genes and functions of osa-miR396d
[0052]
[0053] The target of osa-miR396d is the growth-regulating factor gene Os06...
Embodiment 3
[0054] Example 3. Cutting of target gene mRNA by miRNA
[0055] The cleavage of osa-miR396d to the mRNA of the target gene Os06g02560 (see sequence 3 in the sequence listing) was verified by the 5' RACE method (Jones-Rhoades M W, Bartel D P. Computational identification of plant miRNAs and their targets, including a stress-induced miRNA . Mol. Cell, 2004, 14: 787-799.). After the target gene mRNA is cleaved by miRNA, the relatively stable 3' cleavage product 5' end nucleotide phosphate group is exposed, and the 5' end of the cleavage product is ligated with a 5' RACE special linker with T4 RNA ligase; through reverse transcription reaction Synthesize cDNA; perform the first round of PCR with target gene-specific nested outer primers and the nested outer primers included in the kit, and perform the second round with target gene-specific nested inner primers and the nested inner primers included in the kit PCR;
[0056] The precise target gene mRNA cleavage site can be known b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com