Vibrio cholera analysis typing kit
A Vibrio cholerae and kit technology, applied in the field of Vibrio cholerae molecular typing detection kits, can solve problems such as misjudgment and overlap of amplified products, and achieve the effect of simple experimental operation and simple typing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
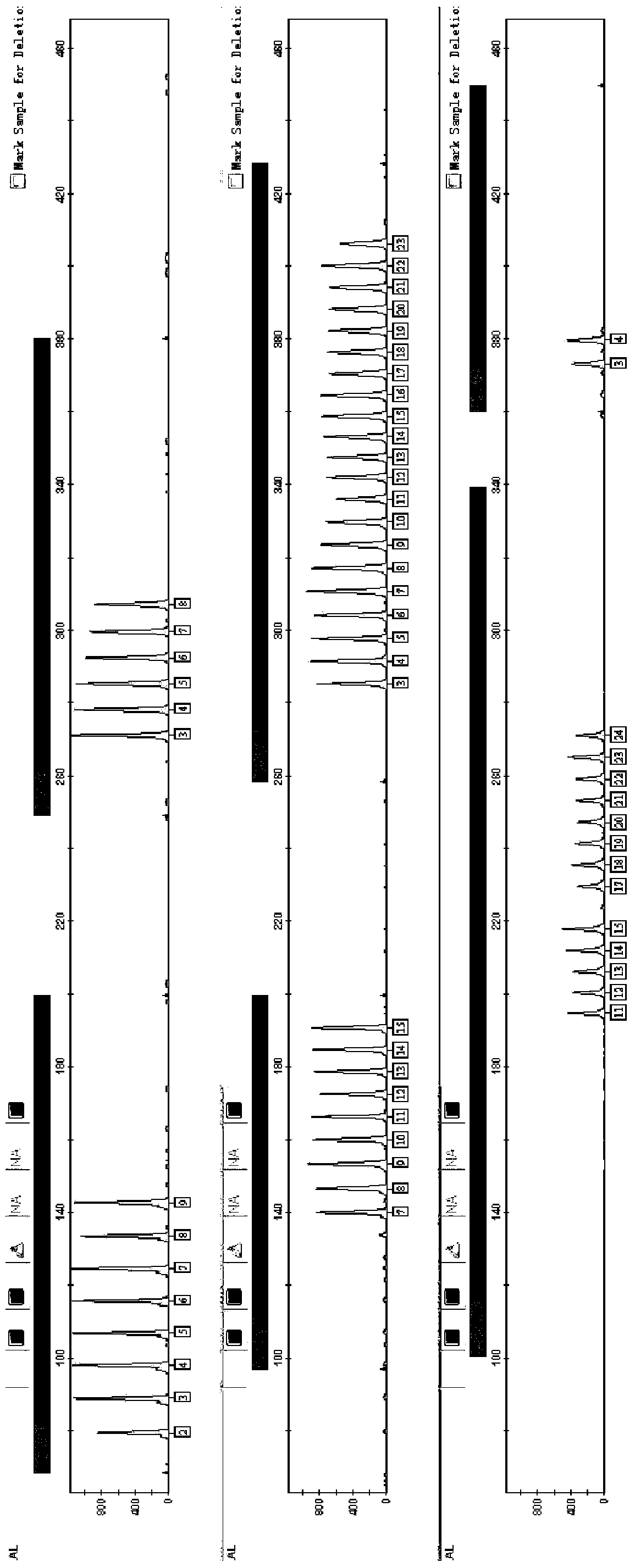
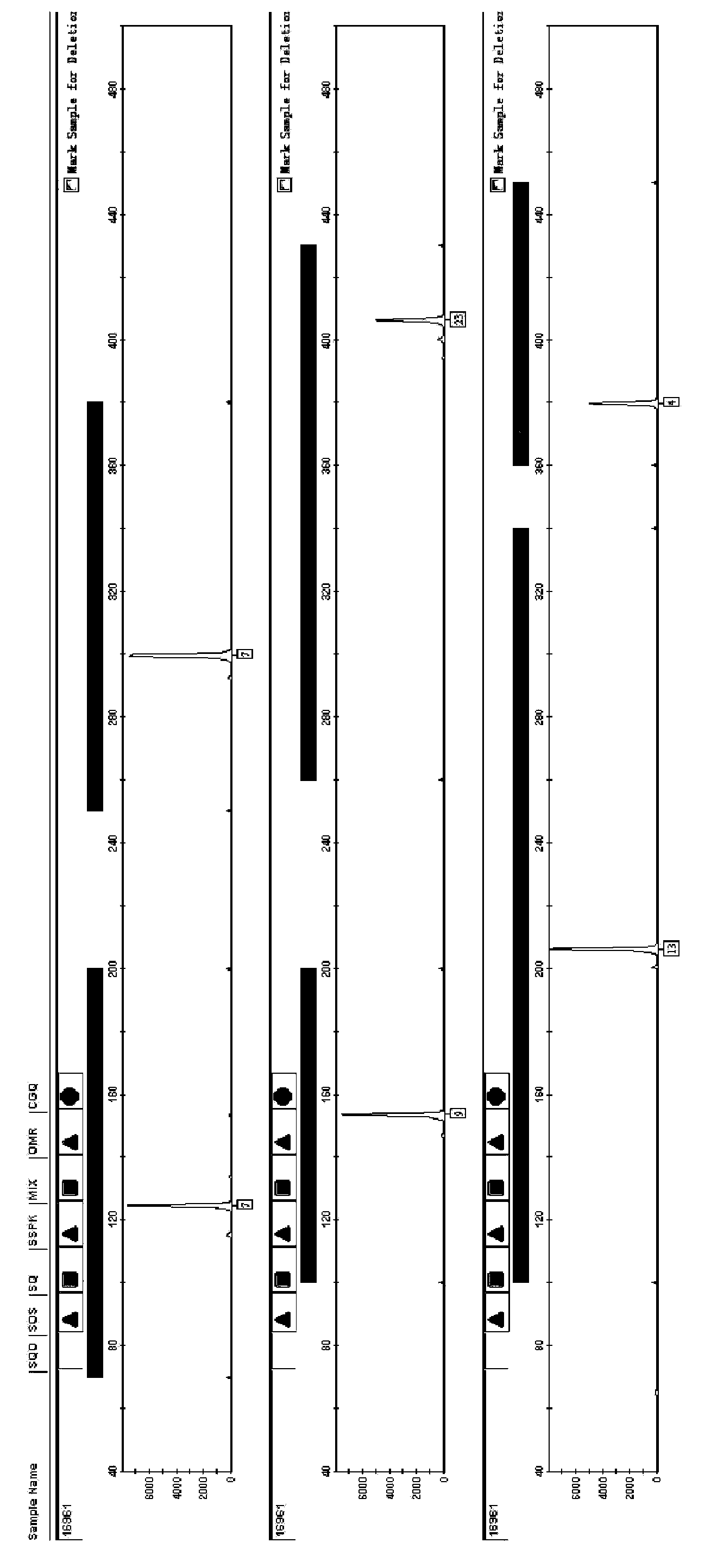
Image
Examples
Embodiment 1
[0029] The design of embodiment 1 kit and the determination of amplification conditions
[0030] 1. Selection of loci
[0031] Six VNTR loci of V. cholerae with high polymorphism were selected, and the relevant information of their alleles is shown in Table 3. The template sequence was downloaded, the locus sequence was downloaded from NCBI genebank, and the position of the sequence repeat region was determined. Allelic ranges were determined by aligning the sequences of different strains of V. cholerae at the same locus in Genebank. The result is as follows:
[0032] Table 4 loci and allele information
[0033] loci
Serial Genebank number
repeat location
allele range
VC1650
CP002555.1
1646667-1646729
2-14
VC0437
CP003330.1
466934-467328
2-24
VC0147
CP003330.1
137105-137159
2-17
VCA0171
CP003331.1
187738-187875
6-26
VCA0283
CP003331.1
303918-303989
3-30
...
Embodiment 2
[0085] The preparation of embodiment 2 allelic typing standards
[0086] The Vibrio cholerae strains collected by the Zhejiang Provincial Center for Disease Control and Prevention were tested, and samples of the same locus and alleles of different sizes were selected. The following table shows the composition of the alleles of each locus:
[0087] Table 14 Allele Information
[0088]
[0089] Amplify with VC1650, VC0437, VC0147, VCA0171, VCA0283 and VC1457 single locus amplification primers (primer concentration 10 μM) respectively. The genomic DNA sample can be bacterial liquid or extracted DNA.
[0090] The primer sequences are as follows:
[0091] Table 15 Primer Sequence
[0092]
[0093] A. The amplification system is as follows:
[0094] Table 16 Kit Configuration Composition
[0095]
[0096] The Reaction Mix mentioned in it is MgCl 2 7.5mM, Tris-HCl buffer125mM, KCl125mM, dNTPs7.5mM, BSA2mg / ml.
[0097] B. Amplification thermal cycle
[0098] Table 17 ...
Embodiment 3
[0108] Embodiment 3: MLVA detection of 19 bacterial strains
[0109]The strains were derived from the Vibrio cholerae strains preserved by the Zhejiang Provincial Center for Disease Control and Prevention, and 19 strains collected at different times were randomly selected. These samples were classified into 12 types by the CDC using the classic PFGE method. Nos. 10-14 belonged to the same type, Nos. 15, 16, 17, and 19 belonged to the same type, and the other strains were the only type.
[0110] By the experimental system of embodiment 1, amplification procedure, detection method detects, and 19 strains typing results are as follows:
[0111] Table 18 strain typing results
[0112]
[0113] It can be seen from the results in the table that the 19 strains were divided into 17 types, among which the experimental numbers 12 and 13, and 17 and 19 were respectively the same type, showing better typing ability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com