Method for rapidly accurately discriminating crassostrea rivularis and crassostrea gigas
A technology for Pacific oysters and near river oysters, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of difficult distinction and inaccurate classification of Pacific oysters and near river oysters, and achieve the effect of low cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
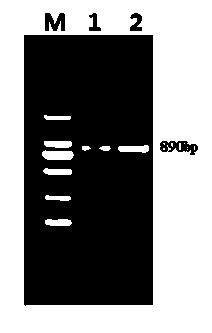
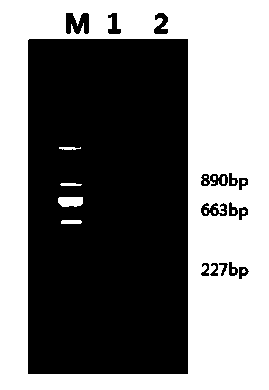
[0017] a. Two kinds of oysters were selected (the Pacific oyster culture group was purchased from Tianjin Aquaculture Farm, and the nearby oyster samples were purchased from Hainan Aquaculture Farm), and the adductor muscle of each oyster was placed in EP tubes, and 70% ethanol was added, -20 Store at ℃ for later use; refer to the classic phenol-imitation extraction method of Sambrook et al. to extract the whole genome DNA of oysters from the frozen oyster adductor muscle, and dissolve the DNA samples in deionized sterilized water at a concentration of 100ng / ml. Freeze at -20°C for later use;
[0018] b. Design a primer sequence suitable for the amplification of the target fragment of the two oyster CaM genes, perform PCR amplification on the sample gene obtained in step a, and obtain the PCR product of the target fragment;
[0019] Upstream primer CaM F: 5' ATGGCCGATCAACTCACAG 3'
[0020] Downstream primer CaM R: 5' GCCTTGAGTGGAAGTCGAT 3'
[0021] c. The obtained PCR pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com