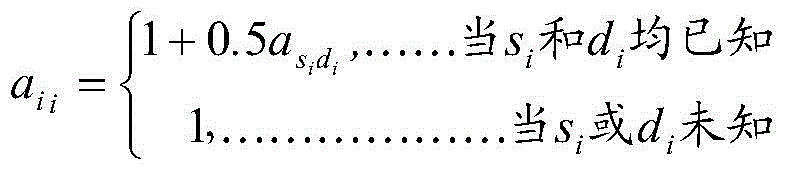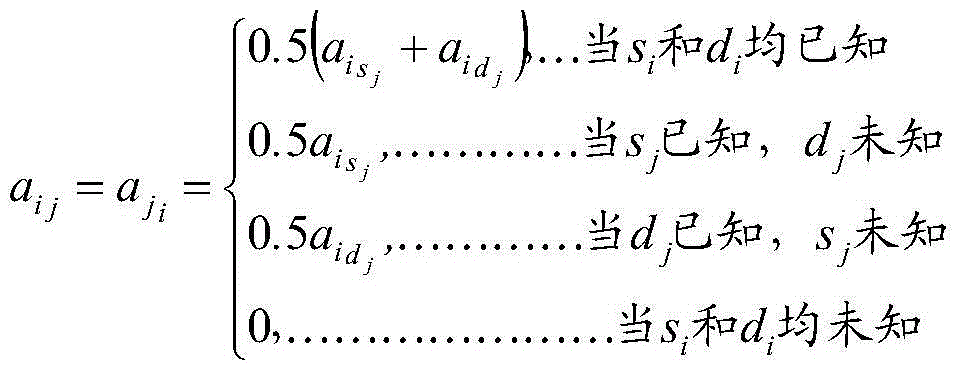Method for rapidly evaluating genome breeding value and application
A genome and breeding value technology, applied in the field of bioinformatics, can solve problems such as exponential increase of calculation time, affecting the efficiency of genome breeding value evaluation, implementation difficulties, etc., and achieve the effect of speeding up calculation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
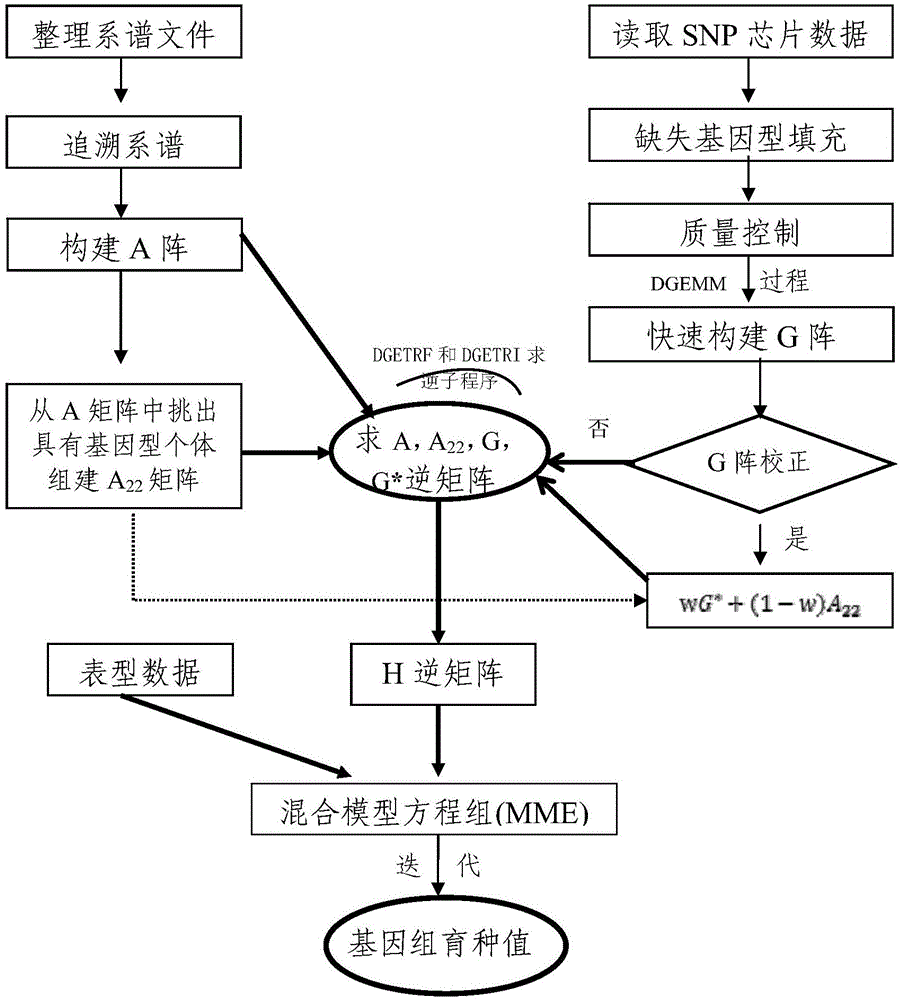
[0029] see figure 1 , this embodiment provides a method for quickly estimating the genome breeding value, comprising the following steps,
[0030] 1) Obtaining data, the data comes from 5974 Chinese Holstein cows, born between 2004 and 2012, all cows were tested for Illunima50K SNP chip genotype, and the milk yield and milk protein amount of 5 milk production traits , milk protein rate, milk fat amount, and milk fat rate were estimated by traditional breeding values. And edit the SNP chip data, a total of 47,160 SNPs were used for analysis; according to the pedigree information of 5,974 cows with SNP genotypes, the data of 10 generations were traced upwards, including 130,852 individuals, which were used for the construction of the kinship matrix A array; the reliability of traditional breeding values was selected The breeding values of 5 milk production traits of individuals higher than 0.45 were used as phenotypes to generate the final phenotype data files.
[0031] 2)...
Embodiment 2
[0056] 7) Repeat step 6) to estimate the genomic breeding values of other 4 milk production traits such as milk protein amount. Embodiment 2 uses real data to evaluate the calculation speed and accuracy of this method
[0057] Using the data in Example 1, use the present invention to calculate and construct the H matrix and the inversion method, and compare the calculation time with the traditional symmetric matrix inversion and the R language ginv function. At the same time, compared with the traditional GBLUP method that only uses genome information, the accuracy of predicting genome breeding value is compared. In Example 1, 541 cows born after 2008 and 2008 with 5974 SNP genotypes were selected, and their traditional breeding value was correlated with the genome breeding value as a standard to measure the accuracy of genome breeding value prediction. The higher the correlation, the higher the explanation The more accurate the genome breeding value is estimated, the bette...
Embodiment 3
[0067] Embodiment 3 simulation data
[0068] The simulation data is an outbred population simulated by QTL-MAS Workshop using LDSO software in 2011. It is constructed by simulating 1000 generations of historical groups with 1000 individuals per generation, and 30 generations of contemporary groups with 150 individuals per generation. The data used in the analysis is the last generation data, a total of 20 male animal lines, each male animal mated with 100 female animals, and each female animal produced 15 offspring. 9990 SNP markers are evenly distributed on 5 chromosomes, each chromosome is 1 Morgen in length, and 1998 SNP markers are evenly distributed.
[0069] The specific data information provided by the simulation data are: pedigree files, SNP marker files, and phenotype files, where the phenotype files are the phenotype values of 10 individuals in each 15 full-sibling offspring, and the other 5 individuals to verify the individual. Also provides the true breeding va...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com