Immunosensor based on hybrid chain reaction and single molecule counting and application of immunosensor
A hybridization chain reaction and single-molecule counting technology, applied in the field of immunosensors, can solve the problems of affecting the authenticity of detection, prone to false positives, and poor repeatability, so as to reduce the generation of false positive signals and reduce the interference to the background , the effect of high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
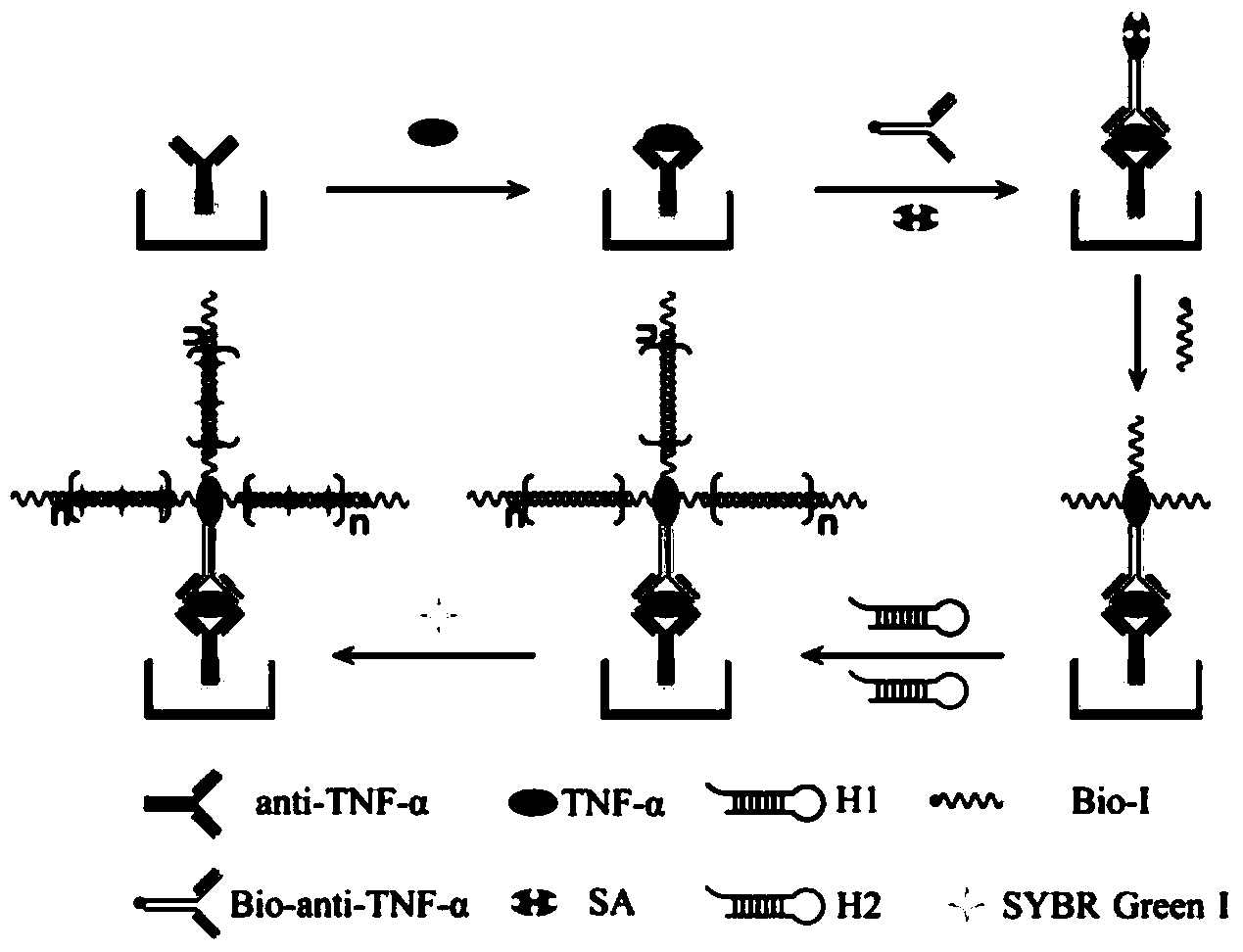
[0056] An immunosensor based on hybridization chain reaction and single molecule counting, composed of tumor necrosis factor-α capture antibody (anti-TNF-α), monoclonal rabbit anti-human tumor necrosis factor-α detection antibody (Bio-anti-TNF-α ), streptavidin, biotinylated primers and hairpin probes;
[0057] The base sequence of the biotinylated primer (Bio-1) is 5'-AGT CTA GGA TTC GGC GTG GGT TAA TTT TTT TTT-biotin-3';
[0058] The hairpin probe consists of a hairpin probe H1 and a hairpin probe H2, wherein the base sequence of the hairpin probe H1 is 5'-TTA ACC CAC GCC GAA TCC TAG ACT CAA AGT AGT CTA GGA TTC GGC GTG-3';
[0059] The base sequence of hairpin probe H2 is 5'-AGT CTA GGA TTC GGC GTG GGT TAA CAC GCC GAA TCC TAG ACT ACT TTG-3'.
[0060] The preparation method of the immunosensor comprises the following steps:
[0061] (1) Construction of the sandwich immune reaction process: 50 μL of tumor necrosis factor-α capture antibody (anti-TNF-α) (20pM) was dropped ont...
Embodiment 2
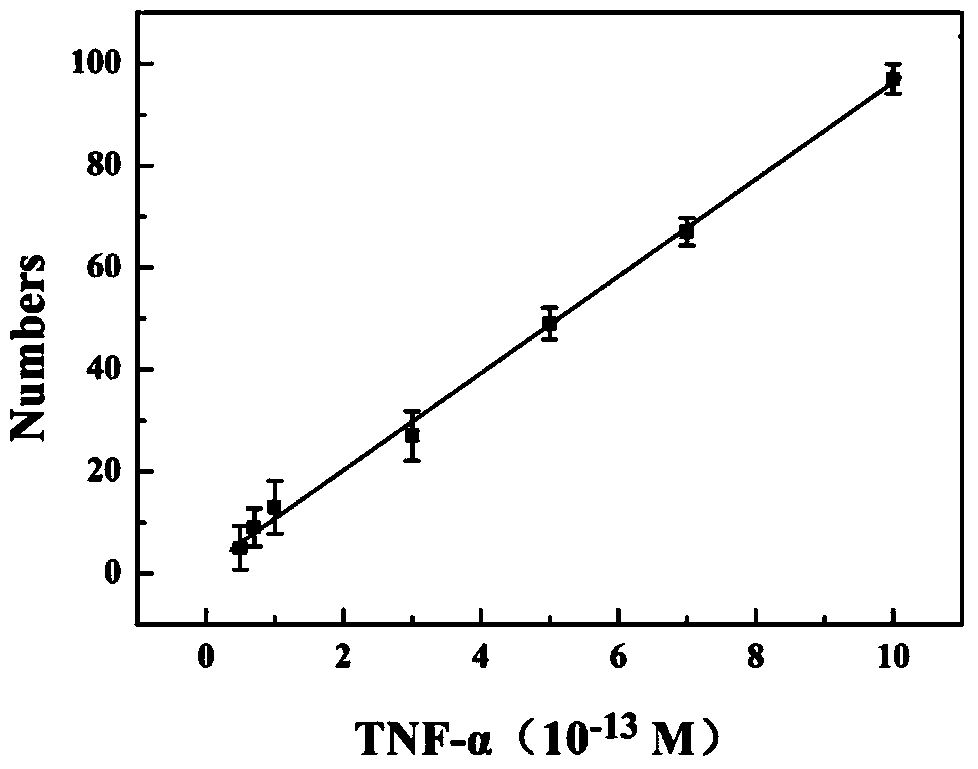
[0072] An immunosensor based on hybridization chain reaction and single molecule counting, in its preparation method, the concentration of anti-TNF-α is 10pM; the blocking agent is bovine serum albumin, and the concentration is 60mM; the concentration of Bio-anti-TNF-α is 20pM, the concentration of streptavidin is 10pM, the concentration of Bio-I is 0.5nM, in the mixture of hairpin probes H1 and H2, the concentrations of H1 and H2 are the same, both are 5nM;
[0073] The volume ratio of the mixture of anti-TNF-α, blocking agent, Bio-anti-TNF-α, streptavidin, Bio-I, hairpin probes H1, H2 is 1:2:1:2:1 ; All the other are with embodiment 1.
Embodiment 3
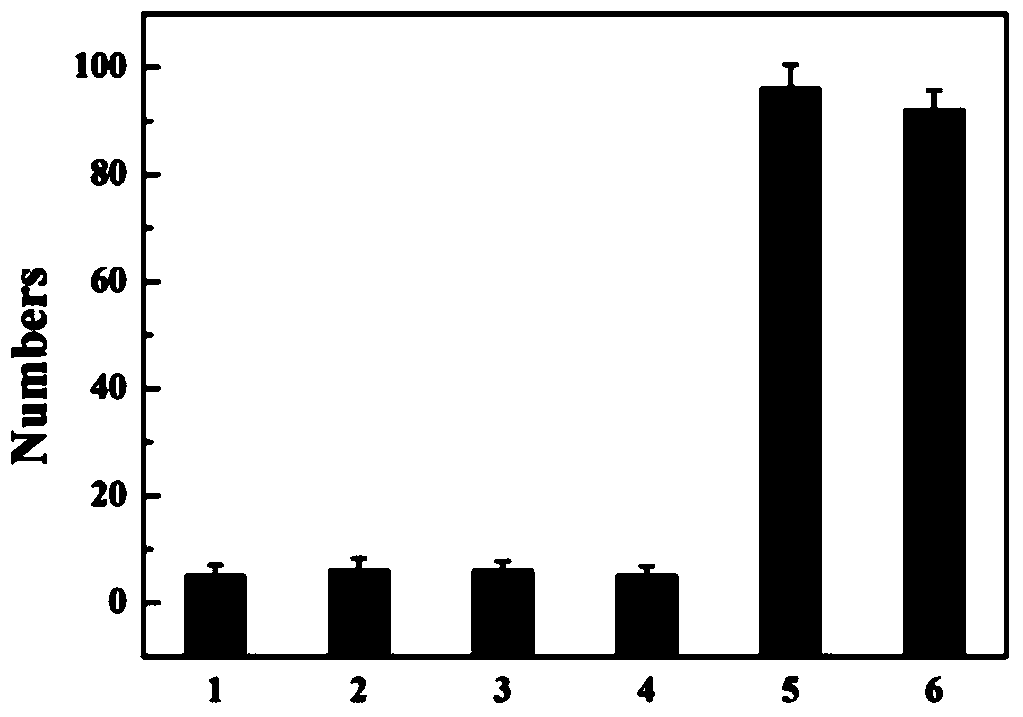
[0075] An immunosensor based on hybridization chain reaction and single molecule counting, in its preparation method, the concentration of anti-TNF-α is 30pM; the blocking agent is bovine serum albumin, and the concentration is 20mM; the concentration of Bio-anti-TNF-α is 40pM, the concentration of streptavidin is 80pM, the concentration of Bio-I is 0.1nM, in the mixture of hairpin probes H1 and H2, the concentrations of H1 and H2 are the same, both are 15nM;
[0076] The volume ratio of the mixture of anti-TNF-α, blocking agent, Bio-anti-TNF-α, streptavidin, Bio-I, hairpin probes H1, H2 is 2:1:2:1:2 ; All the other are with embodiment 1.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com