A multiple PCR primer used for simultaneously detecting five pathogenic bacteria and three virulence genes in marine, and a designing method thereof
A technology for virulence genes and pathogenic bacteria, which is applied in the field of multiplex PCR primers and can solve problems such as loss of bands and loss of target fragments.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0100] Using the primer design software Oligo6.0 to design a multiplex PCR for the simultaneous detection of Enterococcus faecalis, Edwardsiella tarda, Carnobacterium thuringiensis, Vibrio alginolyticus and Enterobacter cloacae gyrB gene, Vibrio parahaemolyticus tlh gene, Enterococcus faecalis cylA gene The primer combination, the formed primer combination contains 23 primer bases, the melting temperature Tm value of the primer is 60°C, and the GC% of the primer is 60%;
[0101] Step 2, screening the primers in the primer combination, retaining primers that do not form primer dimers;
[0102] Step 3, judging the competitive status of the retained primers, comparing the GC% of the above-mentioned retained primers with the number of bases, when the base numbers of the primers are the same and the GC% of the inner primer is less than the GC% of the outer primer, or When the number of bases of the primers is not exactly the same and the base number of the inner primer is one base ...
Embodiment 2
[0123] Example 2 Multiple PCR primer design method for simultaneous detection of Enterococcus faecalis, Edwardsiella tarda, Carnobacterium thuringiensis, Vibrio alginolyticus and Enterobacter cloacae gyrB gene, Vibrio parahaemolyticus tlh gene, Enterococcus faecalis cylA gene , the method includes the following steps:
[0124] Step 1, use the primer design software Primer Premier5.0 to design and simultaneously detect Enterococcus faecalis, Edwardsiella tarda, Carnobacterium thuringiensis, Vibrio alginolyticus and Enterobacter cloacae gyrB gene, Vibrio parahaemolyticus tlh gene, Enterococcus faecalis cylA The multiple PCR primer combination of the gene, the number of primer bases contained in the formed primer combination is 22, the melting temperature Tm value of the primer is 54°C, and the GC% of the primer is 40%;
[0125] Step 2, screening the primers in the primer combination, retaining primers that do not form primer dimers;
[0126] Step 3, judging the competitive stat...
Embodiment 3
[0148] 1. Design and synthesize primers for Enterococcus faecalis, Edwardsiella tarda, Carobacillus, Bacillus thuringiensis and Vibrio alginolyticus, Enterobacter cloacae gyrB gene, Vibrio parahaemolyticus tlh gene, Enterococcus faecalis cylA gene
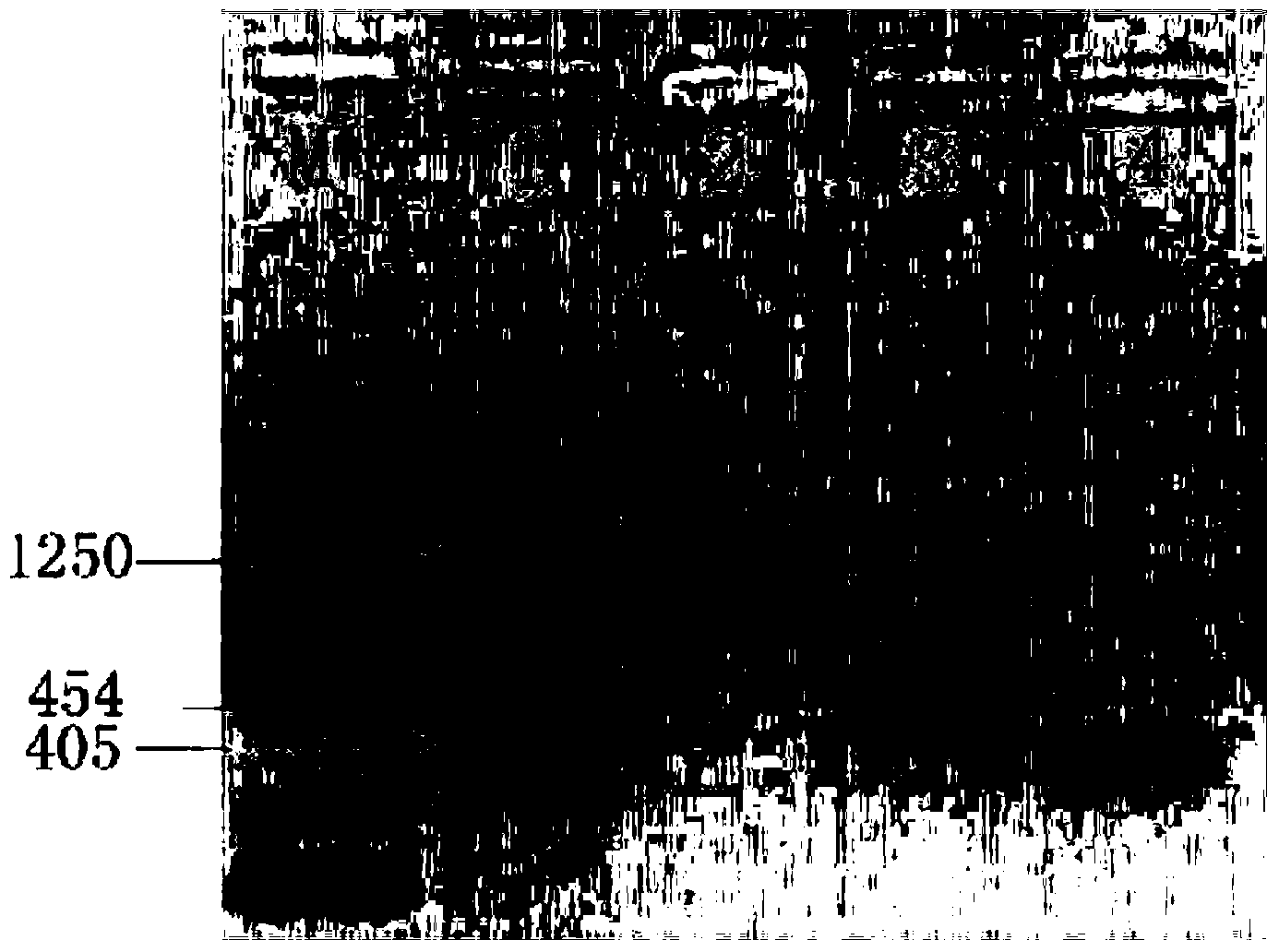
[0149] Referring to the corresponding gene sequences of the marine isolates and the corresponding gene sequences of the four bacteria published on GenBank, a pair of specific primers were designed by using primer express2.0. The size of the PCR amplified fragment corresponding to the primers is 708bp, the size of the PCR amplified fragment corresponding to the Bacillus thuringiensis primer is 566bp, the size of the PCR amplified fragment corresponding to the primer of Bacillus thuringiensis is 458bp, and the size of the PCR amplified fragment corresponding to the primer of Vibrio alginolyticus is 159bp, the size of the PCR amplification fragment corresponding to the primers of the Enterobacter cloacae gyrB gene is 1250bp, the size o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com