Production of isoprene, isoprenoid precursors, and isoprenoids using acetoacetyl-coa synthase
A technology of acetoacetyl coenzyme and acetyl coenzyme, which is applied in the directions of acyltransferase, biochemical equipment and methods, and introduction of foreign genetic material using vectors, can solve the problem of expensive purification, uncommercially attractive and time-consuming isoprene yield. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
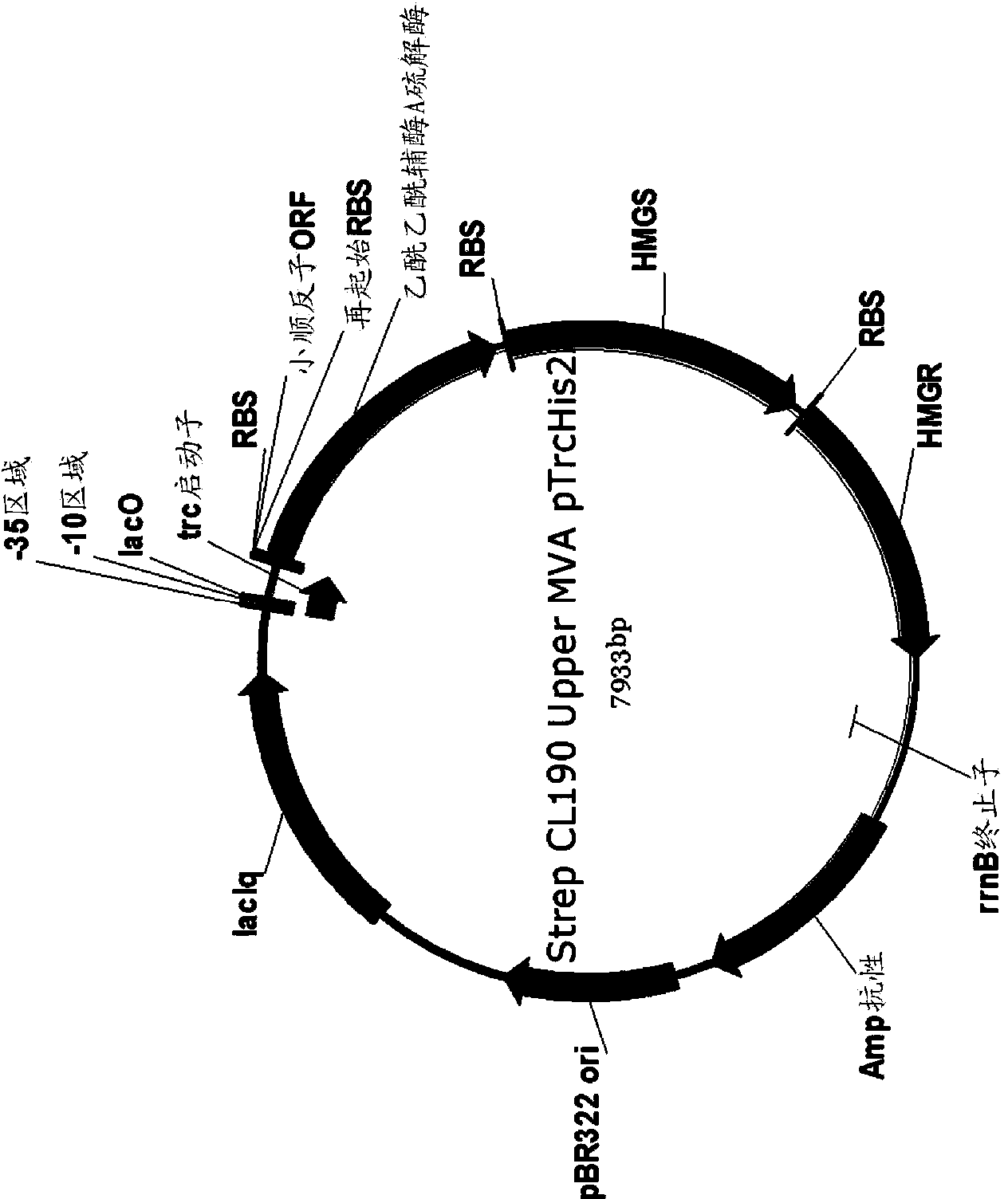
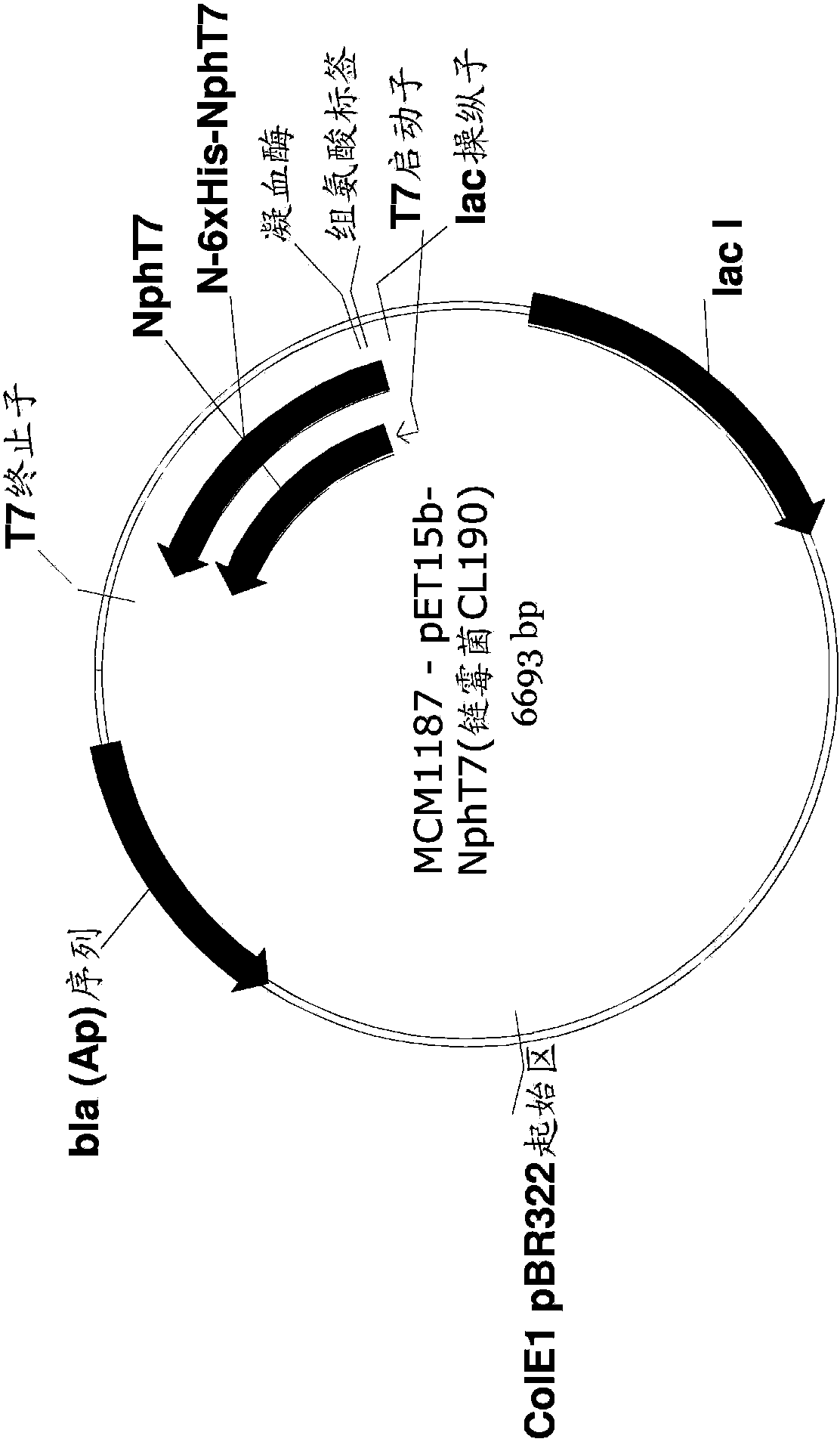
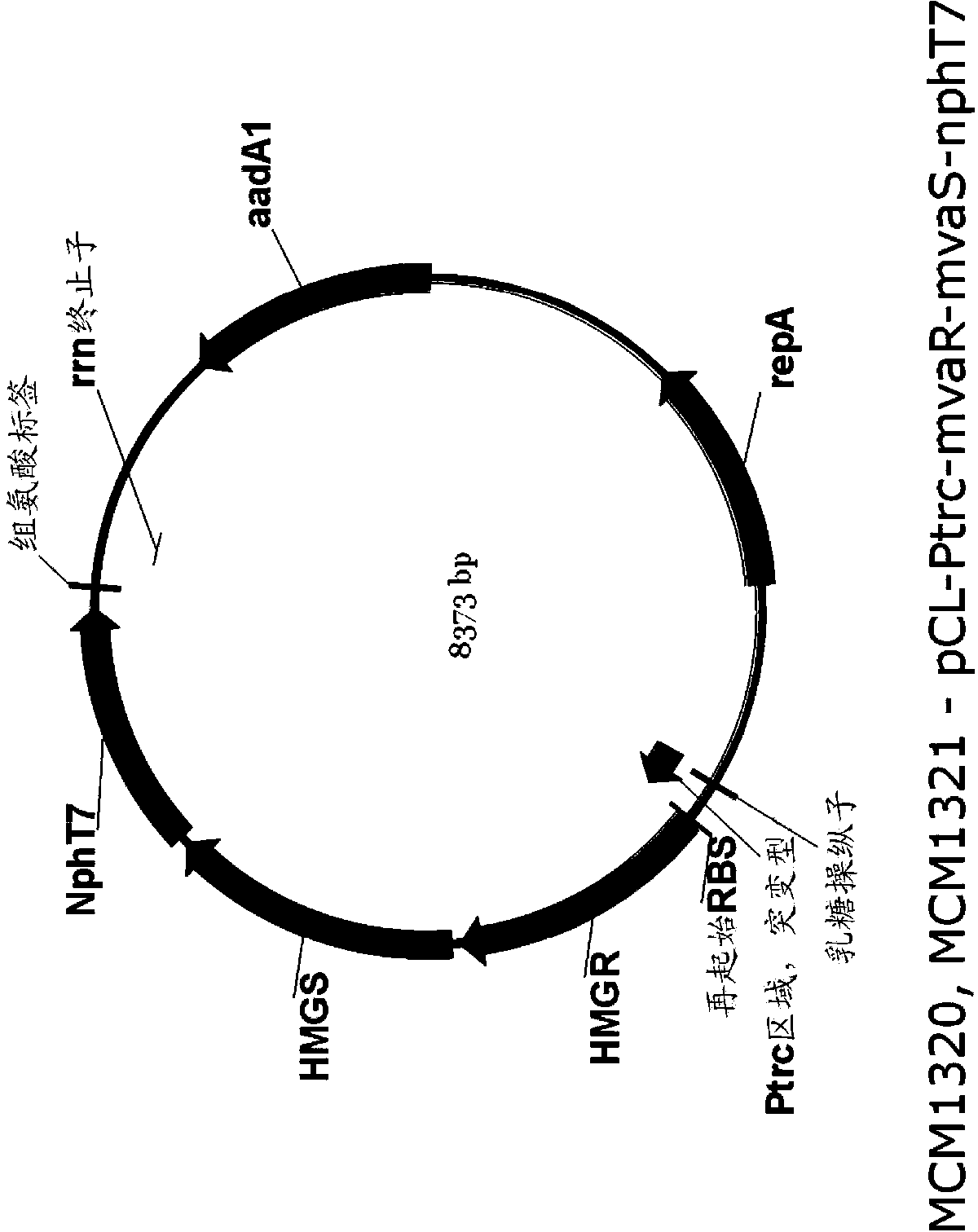
[0259] Example 1: Encoding MVA for the production of MVA by acetoacetyl-CoA synthase (NphT7) Construction of Upstream Pathway Plasmid
[0260] An expression plasmid was generated to encode the nphT7 gene, mvaS gene, and mvaR gene for expressing acetoacetyl-CoA synthase, HMG-CoA synthase, and HMG-CoA reductase, respectively. Simply put, forward and reverse primers are synthesized to amplify mvaS genes (MCM489 and MCM490), mvaR genes (MCM491 and MCM492) and nphT7 genes (MCM495 and MCM496) from synthetic genes encoding Streptomyces proteins (Table 1) . The MCM485 forward primer and MCM486 reverse primer were used to amplify the expression vector. The DNA template used for vector amplification is pMCM1225 (Table 2). The DNA template used to amplify mvaS and mvaR from Streptomyces is StrepCL190 (DNA2.0), which contains synthetic operons encoding mvaS and mvaR, and also encodes acetyl-CoA acetyltransferase (atoB). The pMCM1187 template including the synthetic gene encoding histidi...
example 2
[0273] Example 2: Construction of a plasmid encoding isoprene synthase and MVK for isoprene production
[0274] An expression plasmid for isoprene synthase and mevalonate kinase (MVK) with the bla gene encoding β-lactamase was generated. In short, primers MCM694 and MCM695 (Table 5) were used to amplify the bla gene from pUC19DNA (Invitrogen). The primers MCM696 and MCM697 were used to amplify the expression plasmid pDu65, which did not contain the cmR marker gene. use Seamless Cloning and Assembly Kit ( Seamless cloning and assembly kit) (#A13288) The amplicons are fused according to the manufacturer's protocol, and the product is then transformed into chemically competent MD09-314 cells. The fusion plasmid was selected on the LB / carb50 plate at 37°C overnight. A single colony was selected, cultured in 5mL LB / carb50 at 37°C and stored at -80°C.
[0275] Table 5. Primers used for pMCM1623 construction
[0276]
example 3
[0277] Example 3: Construction of thiolase-deficient Escherichia coli strain CMP861
[0278] Produce acetyl-CoA acetyltransferase (atoB) deficient strains. In short, the strain JW2218 (Baba et al. 2006. Mol. Syst. Biol. ("Molecular Systems Biology") 2:2006.0008) from the Keio collection (Keio collection) was used as a template, and the primer atoBrecF (5' -GCAATTCCCCTTCTACGCTGGG-3' (SEQ ID NO: 15)) and atoBrecR (5'-CTCGACCTTCACGTTGTTACGCC-3' (SEQ ID NO: 16)), by PCR amplifying a DNA fragment containing the atoB gene disrupted by the kanamycin marker . The polymerase Herculase II Fusion (Agilent, Santa Clara, CA) was used according to the manufacturer's instructions. The obtained PCR product was used in a recombineering reaction (Recombineering Reaction) to integrate the PCR product at the atoB locus of the strain CMP451 according to the recommendation of the manufacturer (Gene Bridges, Heidelberg, Germany). CMP451 is CMP258 with two modifications (see US Patent Application No...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com