A kind of recombinant nuclease and preparation method thereof
A technology for recombining nucleic acid and enzyme genes, applied in the biological field, can solve the problem of product inactivity, achieve high expression, simple expression scheme, and improve quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1 Obtaining of recombinant nuclease gene
[0068] According to the Serratia marcescens (S.marcescens) nuclease gene sequence reported in the literature (GenBank: M19495.1), the gene expression sequence was optimized. The optimized sequence is shown in Sequence Table 1, and 20 complementary oligonucleotides were synthesized. According to the conventional method of molecular cloning, first treat with T4 phage polynucleotide kinase at 37°C for 30 minutes, mix the phosphorylated oligonucleotide fragments in equimolar ratio, denature at 94°C for 5 minutes, immediately anneal at 65°C for 10 minutes, then add T4 ligase , ligated overnight at 16°C to obtain target gene template fragments. Take 4 sterile microcentrifuge tubes and add:
[0069]
[0070] The above mixture was shaken gently to mix well, then briefly centrifuged, and then placed in a 14°C water bath for incubation and connection overnight (12-16h). Take 5 μl of the ligation product and add it to 50 μl E...
Embodiment 2
[0071] Example 2 Expression of N-terminal fusion tag recombinant nuclease gene
[0072] Design a pair of primers. Primers 1 and 2 are shown in Sequence Listing Sequence 2 and Sequence 3 respectively. The primer at the 5' end of the gene has an Nde I restriction site, and the primer at the 3' end has a BamH I restriction site. In 0.2ml PCR Prepare a 25μl reaction system in a microcentrifuge tube:
[0073]
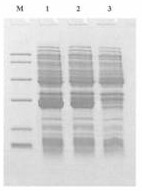
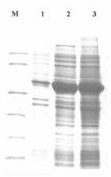
[0074] Pre-denature at 94°C for 5 minutes, set 94°C for 1min, 55°C for 1min, 72°C for 2min, a total of 30 cycles, and finally 72°C for 10min. After PCR, 10 μl of the product was taken for agarose gel electrophoresis, and the fragment size was consistent with the designed size of 777 bp, see sequence 4 in the sequence listing, and sequence 5 for the encoded amino acid sequence.
[0075] The pET-22a plasmid was digested with Nde I and BamH I to recover the large fragment, and ligated with the nuclease gene fragment of PCR. The ratio of the gene fragment to the large vector...
Embodiment 3
[0078] Example 3 Expression of the recombinant nuclease gene with a fusion tag at the C-terminus
[0079] Design a pair of primers, primers 3 and 4 are shown in the sequence table sequence 6 and sequence 7 respectively, the gene 5' end primer has a Nde I restriction site, and the 3' end primer has a BamH I restriction site, in 0.2ml PCR Prepare a 25μl reaction system in a microcentrifuge tube:
[0080]
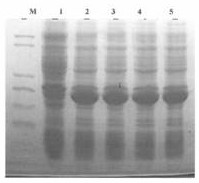
[0081] Pre-denature at 94°C for 5 minutes, set 94°C for 1min, 56°C for 1min, 72°C for 2min, a total of 30 cycles, and finally 72°C for 10min. After the PCR, 10 μl of the product was taken for agarose gel electrophoresis, and the fragment size was consistent with the designed size of 784 bp, see sequence 8 in the sequence listing, and sequence 9 for the encoded amino acid sequence.
[0082] The pET-11b plasmid was digested with Nde I and BamH I to recover the large fragment, and ligated with the nuclease gene fragment of PCR. The ratio of the gene fragment to the large vect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com