Method for establishing DNA (Deoxyribose Nucleic Acid) library based on illumina sequencing platform
A DNA library and construction method technology, which can be applied to libraries, chemical libraries, nucleotide libraries, etc., can solve the problems of limited number of clones, long cycle and high cost, and achieve the effect of shortening construction time and expanding construction scope.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
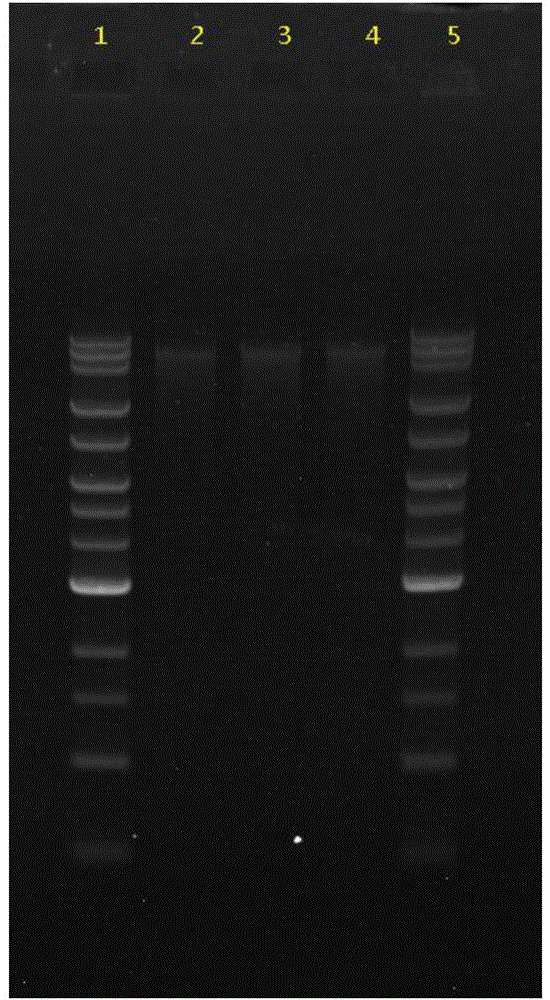
[0056] Example 1 Construction of 20K Large Fragment Library of Turbot Genomic DNA
[0057] 1. Sample DNA interruption:
[0058] First take about 1 ug of sample DNA and break it with a HydroShear large knife head (the speed parameter is set to 15), and use 0.5% agarose gel to detect whether the fragment size is around 20K. If the segment size is not appropriate, adjust the speed code and try again until a suitable interruption condition is selected. The turbot genomic DNA uses this interruption condition, and the DNA fragments are concentrated around 20K, which meets the experimental requirements. Continue to use this condition to interrupt the 20ug DNA sample.
[0059] 2. DNA end filling:
[0060] 1) Prepare the following mixture in a PCR tube:
[0061]
[0062] 2) Gently mix and place at 20°C for 30 min.
[0063] 3) Transfer the DNA into a 1.5ml centrifuge tube, add 200μl Ampure XP beads, mix gently, and place at room temperature for 15min.
[0064] 4) The sample was ...
Embodiment 2
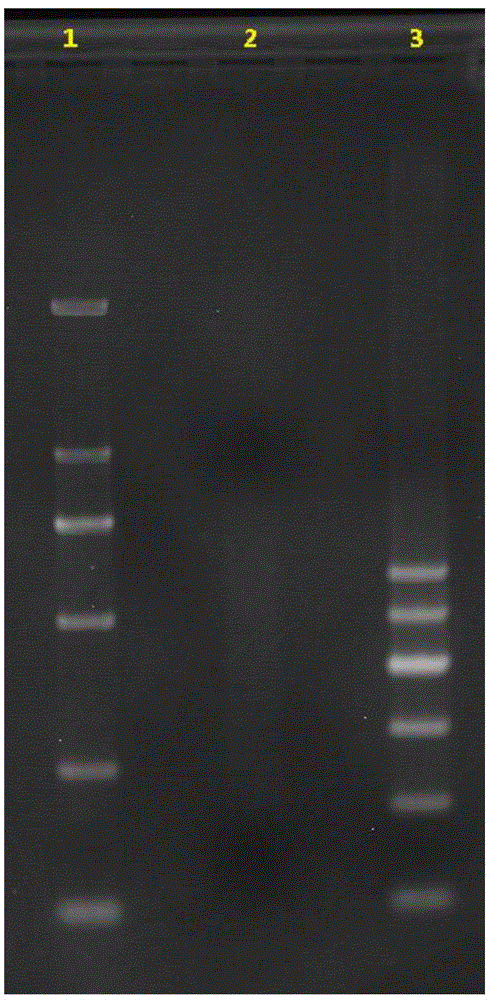
[0170] Example 2 Evaluation of the MP library of the turbot genomic DNA 20K large fragment library
[0171] MP library evaluation was performed on the 20K large fragment library of turbot genomic DNA constructed in Example 1 above.
[0172] The evaluation of the MP library mainly has two important indicators: the average library length that matches the library construction expectation and the ratio of the read pairs with the insert size in the normal range (ie, the circularization rate), which are two important factors to determine the success of the experiment. standard.
[0173] MP library evaluation process: Deduplicate and quality control the sequencing read pairs, then use the software Bowtie2 to compare to the assembled scaffold sequence, use the script written by yourself to compare and compare the results for data statistics, and obtain the statistical values in Table 1:
[0174] Table 1 Evaluation results of 20K-MP library
[0175]
[0176]
[0177] Note: Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com