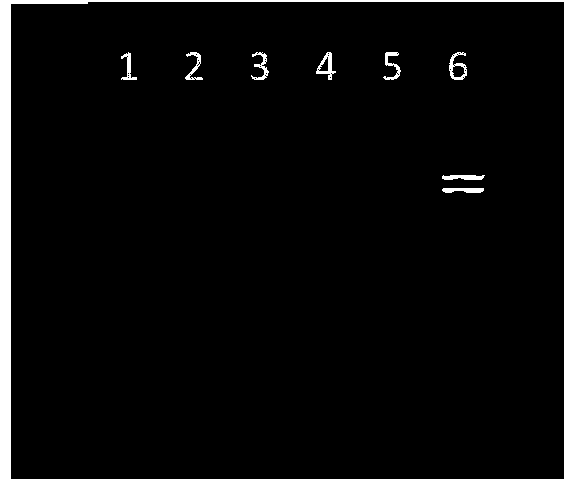Composition for detecting four common drug-resistant genes of escherichia coli
A drug-resistant gene and Escherichia coli technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of cumbersome operation, pseudo-sensitivity, and pseudo-resistance, and save labor and financial resources, good detection effect, and short detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] (1) Primer design, synthesis and kit assembly:
[0071] bla CTX-M-1 、bla CTX-M-9 、bla OXA , aadA designed primers, and the determined primer sequences and amplified fragment lengths for detection are shown in Table 2:
[0072] Table 2
[0073]
[0074]
[0075] On this basis, a kit for mPCR was designed. The kit includes Taq DNA polymerase and PCR reaction solution at a concentration of 5U / μL; the PCR reaction solution contains 10mM Tris HCl, 50mM KCl, 25mM MgCl 2 2.5 mM each of dNTP (dATP, dGTP, dCTP and dTTP) and the above 4 pairs of primers for drug-resistant Escherichia coli (concentrations are shown in Table 2).
[0076] (2) mPCR:
[0077] Use the multiple primers designed in the above steps (1) and corresponding kits to carry out PCR amplification, including the following steps:
[0078] ①Preparation of the sample to be tested: the DNA genome of the sample to be tested is prepared by the kit extraction method:
[0079] a. Food samples:
[0080] Take...
Embodiment 2
[0106] Embodiment 2. Specificity test
[0107] Separately extract Staphylococcus aureus, Listeria monocytogenes, Salmonella typhimurium, Enterococcus faecalis, Escherichia coli (ATCC25922 sensitive strain), Escherichia coli producing CTX-M-1 type ESBLs, and producing CTX-M-9 type ESBLs Escherichia coli, produce OXA type ESBLs Escherichia coli, carry the aminoglycoside antibiotic-resistant Escherichia coli (see Table 1) DNA of aadA gene, according to the detection method established in embodiment 1, get and produce CTX-M-1 type ESBLs Escherichia coli , CTX-M-9 ESBLs-producing Escherichia coli, OXA-producing ESBLs Escherichia coli, and aminoglycoside antibiotic-resistant Escherichia coli DNA templates carrying the aadA gene were each 1 μL as Escherichia coli positive DNA templates, and 1 μL of Staphylococcus aureus, The DNA templates of Listeria monocytogenes, Salmonella typhimurium, Enterococcus faecalis, and Escherichia coli (ATCC25922 sensitive strain) were amplified by mPCR,...
Embodiment 3
[0109] Embodiment 3 detection sensitivity test
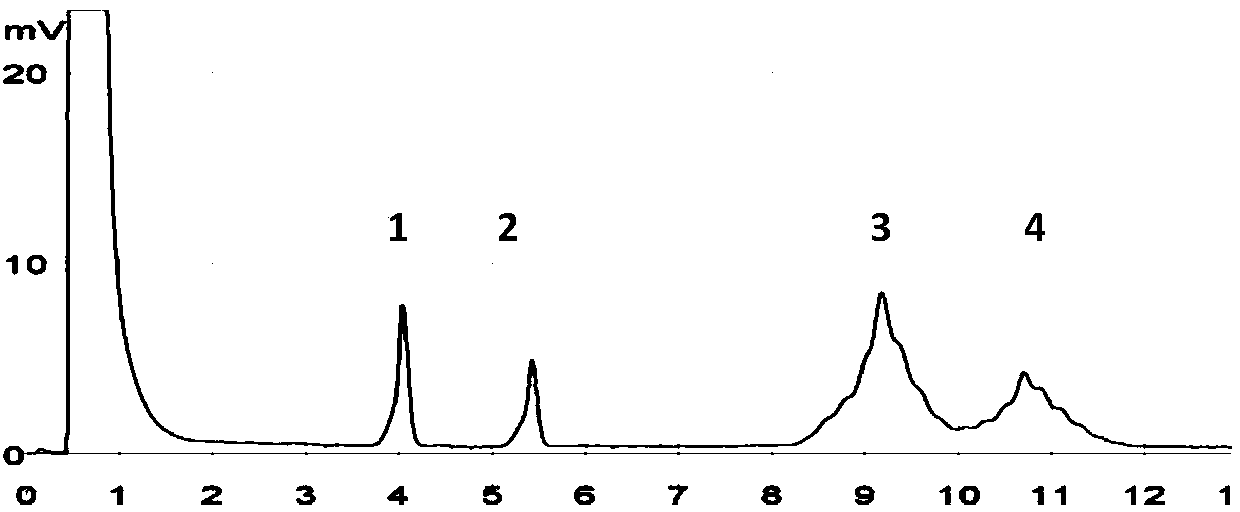
[0110] Inoculate Escherichia coli positive drug-resistant strains (5-5 in Table 1) into TSB medium, culture at 36±1°C for 24 hours, and measure the number of bacteria in the enrichment solution by gradient dilution method or counting method to be 5.4×10 8 cfu / mL, and carry out 10 -1 、10 -2 、10 -3 、10 -4 、10 -5 、10 -6 、10 -7 The serial dilution of the obtained bacterial count was 5.4×10 7 cfu / ml, 5.4×10 6 cfu / ml, 5.4×10 5 cfu / ml, 5.4×10 4 cfu / ml, 5.4×10 3 cfu / ml, 5.4×10 2 cfu / ml, 5.4 × 10cfu / ml bacterial solution, and according to the method established in Example 1 respectively for 5.4 × 10 6 cfu / ml, 5.4×10 5 cfu / ml, 5.4×10 4 cfu / ml, 5.4×10 3 cfu / ml, 5.4×10 2 cfu / ml, 5.4×10cfu / ml bacteria liquid was extracted and detected by electrophoresis, the results are as attached Figure 5 shown. Figure 5 Display: The lowest detection limit of mPCR-electrophoresis detection is 5.4×10 3 cfu / ml.
[0111] According to the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com