SRAP molecule labeled primer for Megalobrama amblycephala family identification, method thereof, and application of primer
A technology of molecular markers and bream, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of complex operation, long time-consuming, difficult application, etc., and achieve simple operation and cost saving , easy-to-sequence effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: Construction of the family sample of bream
[0029] The samples of the 8 bream families were all purchased from the bream breeding base of Huazhong Agricultural University in Ezhou City, Hubei Province. There were 5 male parents and 8 female parents. The parents of the bream families were all from the natural population of Liangzihu. The parent fishes were paired with males and females and artificially propagated, and the hybrid families were established by hybridization. The offspring were numbered L9, L29, L30, L32, L34, L36, L37, and L38. The family information is shown in Table 2. The fins used in the experiment were taken from a small part of the caudal fin of bream and fixed with 95% ethanol.
[0030] Table 2 Information of 8 families of bream used in the experiment
[0031]
Embodiment 2
[0032] Embodiment 2: the screening of primer combination
[0033] The DNA was extracted by conventional methods and diluted to 20 ng / μL, and the most suitable primer pair was selected from the 88 pairs of SRAP primers in Table 1 of the manual. The total volume of the SRAP-PCR reaction system is 15 μL, including TaqDNA polymerase 0.6U (MBI), 10×Buffer 1.5 μL, 2.0 mmol / L Mg2+, 0.25 mmol / LdNTPs (Roche), each primer 0.5 μmol / L, template DNA About 20ng.
[0034] The thermal cycle parameters of PCR were: 94°C pre-denaturation for 5 min; the first 5 cycle parameters were denaturation at 94°C for 1 min, annealing at 35°C for 1 min, extension at 72°C for 1 min; the last 35 cycle parameters were denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and 72 Extend for 1 min at ℃; extend for 10 min at 72°C; cool down to 4°C for storage.
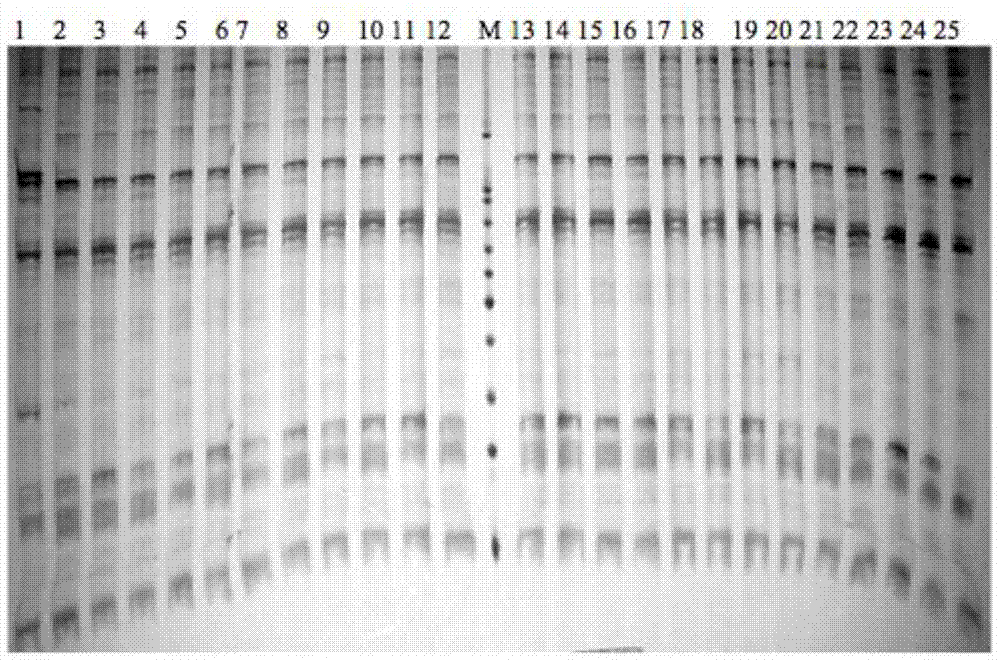
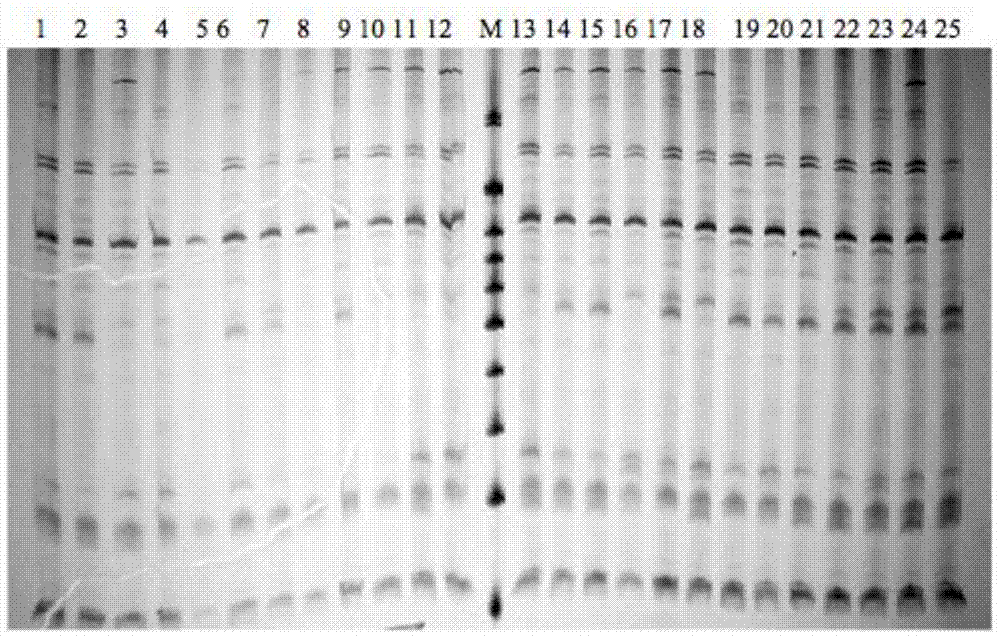
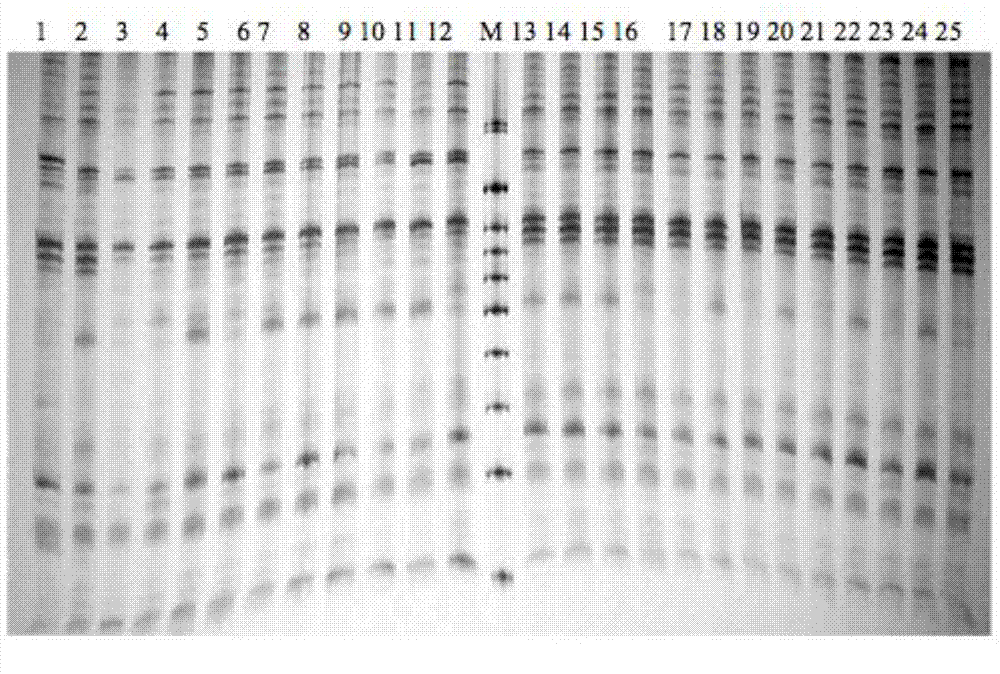
[0035] PCR amplification products were separated by 6% denatured polyacrylamide gel electrophoresis, and the experimental materials used were pr...
Embodiment 3
[0044] Embodiment 3: Genetic diversity and genetic variation analysis of the group head bream population carried out based on the optimal primer combination
[0045] The data processing method is to refer to the 100bp DNA Marker, and count the bands of the obtained SRAP electrophoresis pattern. Each band represents a site, and the band is clearly counted as 1, and no band is counted as 0. A 0, 1 Composed of digital matrix. The POPGENE1.32 program (Yeh et al., 1999) was used to analyze the Nei's gene diversity (h) and Shannon's information index (I) of the 8 families of bream, and calculate the genetic differentiation coefficient among the families (GST), estimated gene flow (Nm) between families based on genetic differentiation coefficients. Molecular analysis of variance (AMOVA) was performed on different families with ARLEQUIN3.11 (Excoffier et al., 2006).
[0046] Using NTSYS-pc2.1 (Rohlf, 2000) to calculate the genetic similarity Dice coefficient (Dice, 1945) of 393 brea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com