Method for digitally and quantitatively detecting nucleic acid based on DNA (deoxyribonucleic acid) chip
A DNA chip and quantitative detection technology, which is applied in the field of bioengineering, can solve problems such as difficult to achieve digital accurate detection level, and achieve the effects of saving amplification time, high feasibility, and high specificity of amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0021] Example 1 Fluorescence probe hybridization method to detect the expression of pain-related gene BDNF in VTA region
[0022] Design the following sequence:
[0023] Bdnf-P: 5'P-GCGTGCAAATTG CCCTATAGTGAGTCGTATTACTATAGTGTCACCGGCAAAGGATCG3';
[0024] Gapdh-P: 5'P GTTATCGTGTAGCCCTATAGTGAGTCGTATTAGTTAGTCACTACTGCACAACTGGTT3';
[0025] IM-primer: 5'amino-TTTTTTTTTTTTTAATACGACTCACTATAGGG3';
[0026] Probe1: 5'cy3-CTATAGTGTCACC3';
[0027] Probe2: 5'cy5-GTTAGTCACTACT3';
[0028] T18: TTTTTTTTTTTTTTTTT.
[0029] The total RNA of mouse VTA region was extracted, and the RNA was converted into cDNA using reverse transcription primer T18 and reverse transcription kit. The reverse transcribed cDNA was added to the ligation reaction system, and Bdnf-P and Gapdh-P were mixed together to carry out the ligation reaction to connect Bdnf-P and Gapdh-P into circular single-stranded DNA (the At the same time, an aldehyde group-modified DNA chip was prepared, and the amino-modified primer...
example 2
[0030] Example 2 Microsequencing was used to detect the expression of pain-related genes BDNF, NNAT and internal reference gene GAPDH in VTA region.
[0031] Design the following sequence:
[0032] Bdnf-P: 5'P-GCGTGCAAATTG CCCTATAGTGAGTCGTATTACTATAGTGTCACCGGCAAAGGATCG3';
[0033] Gapdh-P: 5'PGTTATCGTGTAGCCCTATAGTGAGTCGTATTAGTTAGTCACTACTGCACAACTGGTT3';
[0034] Nnat-P: 5'PGCTGCAGGTGTTCCCTATAGTGAGTCGTATTATTGATCCATGAGACTTCCGCGTGCT3';
[0035] IM-primer: 5'amino-TTTTTTTTTTTTTAATACGACTCACTATAGGG3';
[0036] T18:TTTTTTTTTTTTTTTTTT;
[0037] Seq-primer: CCCTATAGTGAGTCGTATTA.
[0038] According to the method of Example 1, Bdnf-P, Gapdh-P and Nnat-P were ligated into a single-stranded DNA loop. After single-molecule solid-phase amplification, the Seq-primer was hybridized with the amplification template, and then the fluorescently labeled cy3- dCTP, cy5-dGTP, fam-dUTP and the corresponding polymerase system were extended together, and the fluorescence signal was observed under a c...
example 3
[0039]Example 3 Microsequencing was used to detect the expression of pain-related genes BDNF, NNAT and internal reference gene GAPDH in the VTA region of mice with different treatments.
[0040] The reagents and methods are the same as those used in Example 2, except that the prepared immobilized IM-primer divides a DNA chip into different regions by a water-blocking pen or other means, and different regions are used to amplify different treated samples.
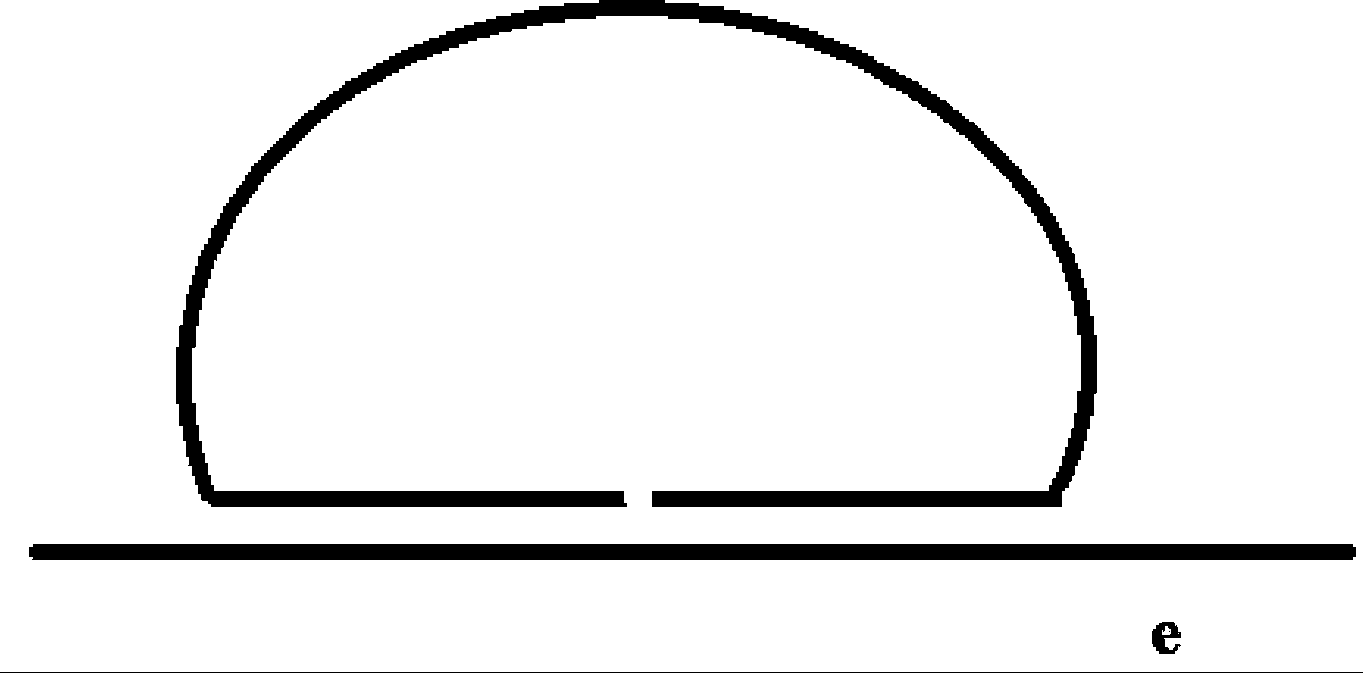
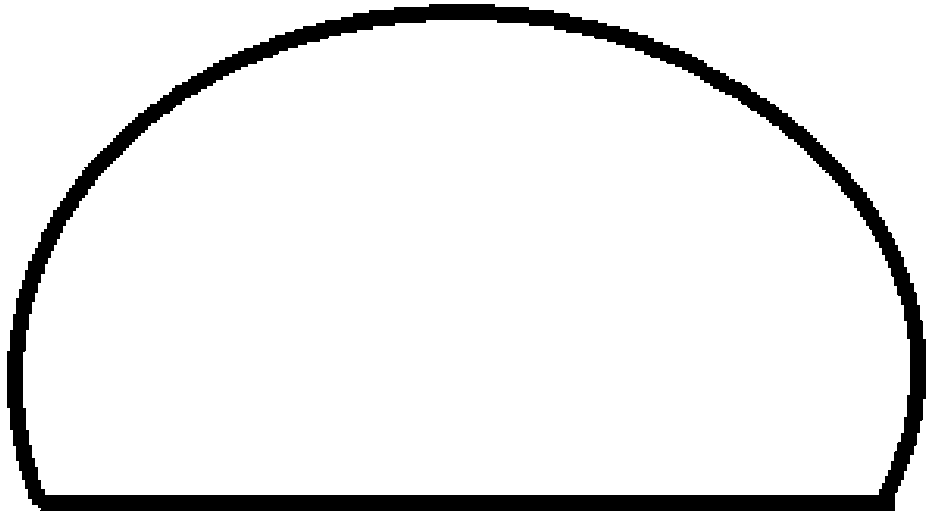
[0041] like figure 1 As shown, the nucleic acid molecule e to be detected, in which a is complementary to d, b is the reverse complementary sequence to the primer sequence immobilized on the DNA chip, c is any DNA sequence, which can be probed by fluorescent labeling through the principle of DNA complementary pairing hybridization Needle detection or detection by micro-sequencing methods, such as figure 2 shown, the ligase system and template e, the sequence can be connected into a loop when the ligase system and template ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com